Dependence of surface aging on DNA topography investigated in attractive bimodal atomic force microscopy
Abstract
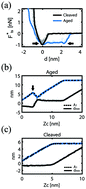
Here we employ bimodal atomic force microscopy (AFM) to investigate the relevance of the aging of the surface and accumulation of adsorbates on the resolved topography of biomolecules. We produce raw bimodal images and a set of contrast channels derived from these to show that the imaging of DNA molecules on hydrophilic model substrates such as mica should be performed immediately after the sample is prepared. Days after preparation, i.e. 48 hours, the adsorbates shield the forces arising from the true substrate and molecule and the molecule might become “invisible” in the images. We employ dsDNA molecules on mica as a model system since the nominal height of dsDNA is comparable to the height of the adsorbed films. With this set up, the molecules can fully disappear under attractive imaging due to the shielding effects of the adsorbates. We further transform the images obtained immediately after cleaving the mica surface and show that the data are then suitable to be transformed into more physically meaningful maps such as Hamaker maps.


 Please wait while we load your content...
Please wait while we load your content...