Structural analyses of isolated cyclic tetrapeptides with varying amino acid residues†
Abstract
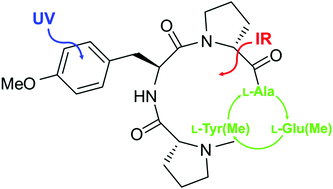
Cyclic peptides represent a large class of substances that occur in nature with important biological and medical functions. Synthetic cyclic peptides are used as artificial receptors due to a series of advantages over conventional receptors. In order to optimize their binding abilities, investigations of their intrinsic structural properties especially with regard to the influence of different amino acid residues are fundamental. Here we report the structural analysis of two synthetic cyclic tetrapeptides cyclo[L-Tyr(Me)-D-Pro-L-Ala-D-Pro] (CPAla) and cyclo[L-Tyr(Me)-D-Pro-L-Glu(Me)-D-Pro] (CPGlu) in a molecular beam by means of combined IR/UV spectroscopic techniques. Structural assignments were achieved by comparing experimentally obtained vibrations and harmonically calculated frequencies including dispersion corrections (B3LYP-D3/TZVP). The investigated cyclic peptides contain an arrangement of an amino acid sequence which is no longer symmetric compared to the former investigations of the cyclo[L-Tyr(Me)-D-Pro]2 peptide. It turns out that all investigated compounds prefer conformations stabilized by two internal hydrogen bonds. In the case of CPGlu containing a flexible side chain with a terminal hydrogen bond acceptor an additional structure was observed in which a hydrogen bond between the terminal carboxylate group and a ring NH group is formed.
- This article is part of the themed collection: Bunsentagung 2017: Physical Chemistry for Life Sciences


 Please wait while we load your content...
Please wait while we load your content...