Solvent effects on ligand binding to a serine protease†
Abstract
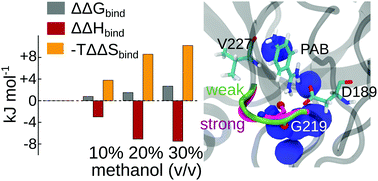
Solvation plays an important role in virtually all biomolecular recognition and binding processes. However, the consequences of changes in solvation conditions often remain elusive. In this work, we combined isothermal titration calorimetry (ITC) and molecular dynamics (MD) simulations to investigate the effect of solvent composition on the thermodynamics of protein–ligand binding. We studied the binding of p-aminobenzamidine (PAB) to trypsin in various water/methanol mixtures as a model system for a biomolecular complex. Our ITC experiments show that the free energy of binding changes only very modestly with methanol concentration, and that this small change is due to strong enthalpy–entropy compensation. The MD and free energy simulations not only reproduce the experimental binding free energies, but also provide atomic-level insights into the mechanisms behind the thermodynamic observations. The more favorable binding enthalpy at increased methanol concentrations (when compared to pure water) is attributed to stronger protein–ligand and intramolecular protein–protein interactions. The stronger protein–ligand interaction is linked to a small-scale conformational rearrangement of the L2 binding pocket loop, which senses the solvent environment. Remarkably, the stronger interactions do not substantially reduce the configurational entropy of the protein. Instead, the more disfavorable entropy contribution to the binding free energy at increased methanol concentrations is due to the desolvation of the ligand from the bulk, which is more favorable in pure aqueous solution than in the presence of methanol. Our work thus underpins the importance of including conformational flexibility, even for an overall rather rigid complex, since even small-amplitude motions can significantly alter the binding energetics. Furthermore, the ability of our combined ITC/MD approach to assign different thermodynamic contributions to distinct conformational states might contribute to an enhanced understanding of biomolecular binding processes in general.
- This article is part of the themed collection: Bunsentagung 2017: Physical Chemistry for Life Sciences


 Please wait while we load your content...
Please wait while we load your content...