Fluorescence-switching RNA for detection of bacterial ribosomes†
Abstract
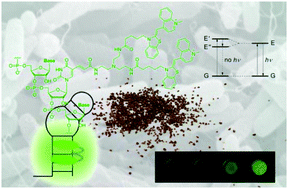
We have developed an efficient chemical system that allows quantification of bacterial ribosomes by fluorescence-based analysis. The key component in the system is the exciton-controlled fluorescent RNA aptamer, which recognizes neomycin B. The intensity of fluorescence from such a ribosome-sensing system increased drastically in the presence of Escherichia coli.

 Please wait while we load your content...
Please wait while we load your content...