Exploring biomolecular energy landscapes
Abstract
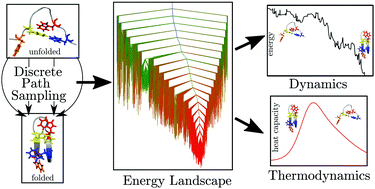
The potential energy landscape perspective provides both a conceptual and a computational framework for predicting, understanding and designing molecular properties. In this Feature Article, we highlight some recent advances that greatly facilitate structure prediction and analysis of global thermodynamics and kinetics in proteins and nucleic acids. The geometry optimisation procedures, on which these calculations are based, can be accelerated significantly using local rigidification of selected degrees of freedom, and through implementations on graphics processing units. Results of progressive local rigidification are first summarised for trpzip1, including a systematic analysis of the heat capacity and rearrangement rates. Benchmarks for all the essential optimisation procedures are then provided for a variety of proteins. Applications are then illustrated from a study of how mutation affects the energy landscape for a coiled-coil protein, and for transitions in helix morphology for a DNA duplex. Both systems exhibit an intrinsically multifunnel landscape, with the potential to act as biomolecular switches.


 Please wait while we load your content...
Please wait while we load your content...