Profiling and quantifying endogenous molecules in single cells using nano-DESI MS†
Abstract
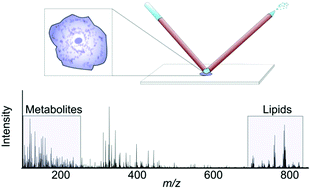
Molecular profiling of single cells has the potential to significantly advance our understanding of cell function and cellular processes of importance to health and disease. In particular, small molecules with rapid turn-over rates can reveal activated metabolic pathways resulting from an altered chemical environment or cellular events such as differentiation. Consequently, techniques for quantitative metabolite detection acquired in a higher throughput manner are needed to characterize the biological variability between seemingly homogenous cells. Here, we show that nanospray desorption electrospray ionization (nano-DESI) mass spectrometry (MS) enables sensitive molecular profiling and quantification of endogenous species in single cells in a higher throughput manner. Specifically, we show a large number of detected amino acids and phospholipids, including plasmalogens, readily detected from single cheek cells. Further, by incorporating a phosphatidylcholine (PC) internal standard into the nano-DESI solvent, we determined the total amount of PC in one cell to be 1.2 pmoles. Finally, we describe a higher throughput approach where molecules in single cells are automatically profiled. These developments in single cell analysis provide a basis for future studies to understand cellular processes related to drug effects, cell differentiation and altered chemical microenvironments.


 Please wait while we load your content...
Please wait while we load your content...