Constraint randomised non-negative factor analysis (CRNNFA): an alternate chemometrics approach for analysing the biochemical data sets†
Abstract
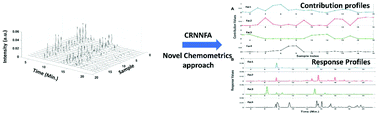
The present work introduces an alternate chemometrics approach constraint randomised non-negative factor analysis (CRNNFA) for analysing the bioanalytical data sets. The CRNNFA algorithm provides the outputs that are easy to interpret and correlate with the real chromatograms. The CRNNFA algorithm achieves termination when the iteration limit is reached circumventing the premature convergence. Theoretical and computational aspects of the proposed method are also described. The analytical and computational potential of CRNNFA are successfully tested by analysing the complex chromatograms of the peptidoglycan samples belonging to the Alphaproteobacterium members. The obtained results clearly show that CRNNFA can easily trace the compositional variability of the peptidoglycan samples. In summary, the proposed method in general can be a potential alternate approach for analysing the data sets obtained from different analytical and clinical fields.


 Please wait while we load your content...
Please wait while we load your content...