RNA model evaluation based on MD simulation of four tRNA analogs†
Abstract
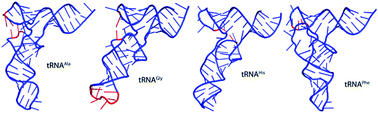
High resolution 3D structures of tRNA molecules are scarce. Therefore, there is a burning need for tools aiming at three-dimensional (3D) RNA structure characterization able to provide accurate information on the tertiary structure of ribonucleic acids. In this study, RNA structure assessment based on primary sequence, molecular dynamics (MD) simulations as well as selected RNA model evaluation methods are used to propose and evaluate four mitochondrial tRNA analogs. The 3D structures of tRNAAla, tRNAGly, tRNAHis, and tRNAPhe are generated from primary sequence using mFold and RNAComposer, and subjected to 100 ns explicit solvent MD simulations with AMBER. The global and local root-mean-square deviations (RMSD), interaction network fidelity (INF), deformation index (DI), the contact area difference-score (CAD-score) as well as principal component analysis (PCA) revealed that the largest changes occurring during MD simulation were in the D–T and anticodon loops but each of the studied tRNA analog is affected differently. tRNAAla and tRNAPhe undergo limited structure perturbation, mostly in the D–T loops regions, while tRNAGly changes the geometry within the anticodon loop. The tRNAHis analog is the most flexible, changes its structure significantly, including separation of the D and T loops. Furthermore, the anticodon loops are visibly more stable than the D–T region and their structure does not change that significantly, except for tRNAGly.

 Please wait while we load your content...
Please wait while we load your content...