Self-assembly of the full-length amyloid Aβ42 protein in dimers†
Abstract
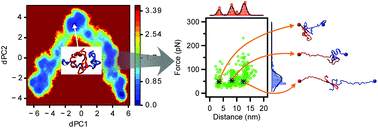
The self-assembly of amyloid (Aβ) proteins into nano-aggregates is a hallmark of Alzheimer's disease (AD) development, yet the mechanism of how disordered monomers assemble into aggregates remains elusive. Here, we applied long-time molecular dynamics simulations to fully characterize the assembly of Aβ42 monomers into dimers. Monomers undergo conformational changes during their interaction, but the resulting dimer structures do not resemble those found in fibril structures. To identify natural conformations of dimers among a set of simulated ones, validation approaches were developed and applied, and a subset of dimer conformations were characterized. These dimers do not contain long β-strands that are usually found in fibrils. The dimers are stabilized primarily by interactions within the central hydrophobic regions and the C-terminal regions, with a contribution from local hydrogen bonding. The dimers are dynamic, as evidenced by the existence of a set of conformations and by the quantitative analyses of the dimer dissociation process.

 Please wait while we load your content...
Please wait while we load your content...