Abstract
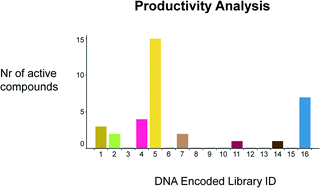
DNA encoded library screens have gained recent interest as they allow for screening of millions of small molecules in a simple manner, with the goal of providing novel chemical starting points in target-based hit identification. Despite this interest, no publication describes the physical properties, novelty, or structural diversity of molecules derived from such screens, nor a comparison of productivity of different DNA encoded libraries. Here we address this gap by analysis of two DNA encoded library screens run against two protein targets employing mixtures of up to 16 different libraries. Fifty-seven exemplar small molecule compounds from 34 structurally distinct clusters were prioritized from the screening results, synthesized and tested for biochemical activity. Thirty-five of the 57 compounds possess significant biochemical activity (IC50 ≤ 10 μM). Seventeen of the 35 biochemically active compounds possess a molecular weight (MW) < 500 Dalton (Da) and clog P < 5, and 6 possess a MW < 400 Da and clog P < 4. None of the 57 DEL-derived compounds exist in the Roche corporate high throughput screening collection and public compound collections. Productivity per library was observed to be independent of library size. The most productive of the 16 investigated libraries was synthesized employing only simple chemistry. Physical properties of DEL-derived compounds correlate with average library properties when truncated sub-libraries are accounted for. Our analysis may help guide the design of future DNA encoded libraries.

 Please wait while we load your content...
Please wait while we load your content...