Combining pseudo dinucleotide composition with the Z curve method to improve the accuracy of predicting DNA elements: a case study in recombination spots†
Abstract
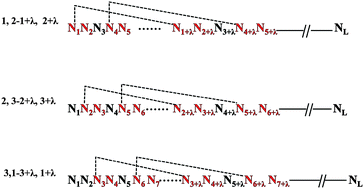
Pseudo dinucleotide composition (PseDNC) and Z curve showed excellent performance in the classification issues of nucleotide sequences in bioinformatics. Inspired by the principle of Z curve theory, we improved PseDNC to give the phase-specific PseDNC (psPseDNC). In this study, we used the prediction of recombination spots as a case to illustrate the capability of psPseDNC and also PseDNC fused with Z curve theory based on a novel machine learning method named large margin distribution machine (LDM). We verified that combining the two widely used approaches could generate better performance compared to only using PseDNC with a support vector machine based (SVM-based) model. The best Mathew's correlation coefficient (MCC) achieved by our LDM-based model was 0.7037 through the rigorous jackknife test and improved by ∼6.6%, ∼3.2%, and ∼2.4% compared with three previous studies. Similarly, the accuracy was improved by 3.2% compared with our previous iRSpot-PseDNC web server through an independent data test. These results demonstrate that the joint use of PseDNC and Z curve enhances performance and can extract more information from a biological sequence. To facilitate research in this area, we constructed a user-friendly web server for predicting hot/cold spots, HcsPredictor, which can be freely accessed from http://cefg.cn/HcsPredictor. In summary, we provided a united algorithm by integrating Z curve with PseDNC. We hope this united algorithm could be extended to other classification issues in DNA elements.

 Please wait while we load your content...
Please wait while we load your content...