Electrochemical DNA biosensors based on long-range electron transfer: investigating the efficiency of a fluidic channel microelectrode compared to an ultramicroelectrode in a two-electrode setup†
Abstract
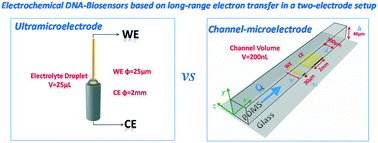
Here, we describe the transposition of an ultramicroelectrode (UME) setup into a microfluidic chip configuration for DNA biosensors. The hydrodynamic properties of the fluidic channel microelectrode were screened with an [Fe(III)(CN)6]3−/[Fe(II)(CN)6]4− redox couple by cyclic voltammetry to provide a basis for further biological processes. A 23-base DNA probe was self-assembled into a monolayer on gold microelectrodes both in classical configuration and integrated in a microfluidic setup. Special interest was focused on the DNA target mimicking the liver-specific micro-ribonucleic acid 122 (miRNA122). Long-range electron transfer was chosen for transducing the hybridization. This direct transduction was indeed significantly enhanced after hybridization due to DNA-duplex π-stacking and the use of redox methylene blue as a DNA intercalator. Quantification of the target was deduced from the resulting electrical signal characterized by cyclic voltammetry. The limit of detection for DNA hybridization was 0.1 fM in stopped flow experiments, where it can reach 1 aM over a 0.5 μL s−1 flow rate, a value 104-fold lower than the one measured with a conventional UME dipped into an electrolyte droplet under the same analytical conditions. An explanation was that forced convection drives more biomolecules to the area of detection even if a balance between the speed of collection and the number of biomolecules collected has been found. The latter point is discussed here along with an attempt to explain why the sensor has reached such an unexpected value for the limit of detection.
- This article is part of the themed collection: Lab on a Chip Recent Open Access Articles

 Please wait while we load your content...
Please wait while we load your content...