Conformational stabilities of iminoallantoin and its base pairs in DNA: implications for mutagenicity†
Abstract
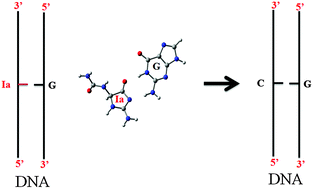
The types of mutations induced by oxidatively damaged products of DNA are continuously in debate. For example, some biochemical studies have proposed that guanidinohydantoin (Gh) would induce exclusively G to C mutations, while other studies have predicted a mixture of various mutations including G to C, G to T and G to A. In addition to the nature of mutations, the exact reasons of these mutations are also not properly understood. It is suggested that Gh can easily isomerize to iminoallantoin (Ia) in a pH-dependent manner and the transition becomes complete at pH > 8. In order to understand Gh/Ia-induced mutations, we have here studied the role of the most stable tautomer of Ia in the R- and S-enantiomeric configurations in promoting mismatch base pair complexes in DNA by employing a density functional theoretical (DFT) approach. It is found that Ia can have 39 different possible tautomeric forms each in the R- and S-enantiomeric configurations, out of which the most stable tautomer would involve the deprotonation of the N1 atom and protonation of the N3 atom. The most stable tautomer of Ia can adopt three different rotameric conformations (Ia1, Ia2, and Ia3) of comparable stabilities. It is further revealed that these rotamers of Ia can interact with different bases of DNA in 88 different possible ways. However, the interaction of G with Ia3 in both the anti- and syn-conformations would be the most stable. It is further revealed that the base pairing patterns, binding energies and electronic environments of anti-Ia3:G and G:T complexes are similar. In addition to this, it is also found that the binding patterns and energies of Gh1:G and Ia3:G complexes are similar. Based on these results, it is proposed that under physiological conditions, Gh1 may be responsible for the observed G to C mutations in DNA, while in an acidic environment Ia3 may be responsible for the same mutations. This study has led to a solid foundation for further high resolution structural studies to completely unravel Ia-induced mutagenicity in DNA.

 Please wait while we load your content...
Please wait while we load your content...