A fast but accurate excitonic simulation of the electronic circular dichroism of nucleic acids: how can it be achieved?†
Abstract
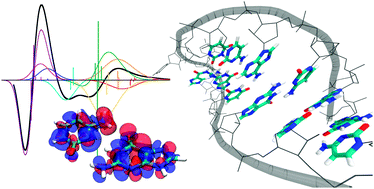
We present and discuss a simple and fast computational approach to the calculation of electronic circular dichroism spectra of nucleic acids. It is based on a exciton model in which the couplings are obtained in terms of the full transition-charge distributions, as resulting from TDDFT methods applied on the individual nucleobases. We validated the method on two systems, a DNA G-quadruplex and a RNA β-hairpin whose solution structures have been accurately determined by means of NMR. We have shown that the different characteristics of composition and structure of the two systems can lead to quite important differences in the dependence of the accuracy of the simulation on the excitonic parameters. The accurate reproduction of the CD spectra together with their interpretation in terms of the excitonic composition suggest that this method may lend itself as a general computational tool to both predict the spectra of hypothetic structures and define clear relationships between structural and ECD properties.

 Please wait while we load your content...
Please wait while we load your content...