A microfluidic-based protein crystallization method in 10 micrometer-sized crystallization space†
Abstract
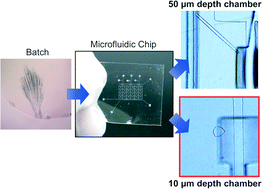
Protein crystallization and subsequent X-ray diffraction analysis of the three-dimensional structure are necessary for elucidation of the biological functions of proteins and effective rational drug design. Therefore, controlling protein crystallization is important to obtain high resolution X-ray diffraction data. Here, a simple microfluidic method using chips with 10 and 50 μm high crystallization chambers to selectively form suitable single protein crystals for X-ray analysis is demonstrated. As proof of concept, three different types of proteins: lysozyme, glucokinase from Pseudoalteromonas sp. AS-131 (PsGK), and NADPH-cytochrome P450 oxidoreductase–heme oxygenase complex were crystallized. We demonstrate that the crystal growth orientation depends on the height of the crystallization chamber regardless of the protein type. Our results suggest that the confined micro space induces the protein molecules to adhere to a specific crystal face and affects the growth kinetics of each crystal face. The present microfluidic-based protein crystallization method can reform a suitable single protein crystal for X-ray analysis from aggregates of needle-shaped protein crystals.


 Please wait while we load your content...
Please wait while we load your content...