Sensitive proton-detected solid-state NMR spectroscopy of large proteins with selective CH3 labelling: application to the 50S ribosome subunit†
Abstract
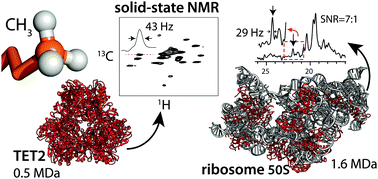
Solid-state NMR spectroscopy allows the characterization of the structure, interactions and dynamics of insoluble and/or very large proteins. Sensitivity and resolution are often major challenges for obtaining atomic-resolution information, in particular for very large protein complexes. Here we show that the use of deuterated, specifically CH3-labelled proteins result in significant sensitivity gains compared to previously employed CHD2 labelling, while line widths increase only marginally. We apply this labelling strategy to a 468 kDa-large dodecameric aminopeptidase, TET2, and the 1.6 MDa-large 50S ribosome subunit of Thermus thermophilus.


 Please wait while we load your content...
Please wait while we load your content...