Mapping activity elements of protegrin antimicrobial peptides by HomoSAR†
Abstract
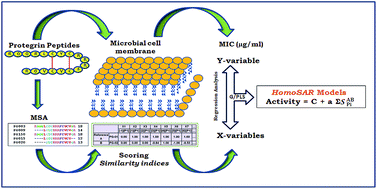
Antimicrobial peptides (AMPs) are naturally occurring small peptides which are an innate part of the host's defense mechanism. They are active against both Gram-negative and Gram-positive bacteria, various viruses, fungi, and parasites. There is little consensus in the amino acid sequences of AMPs but evidently they do possess some definite common features, such as relative hydrophobic and a positively charged amphipathic structure that has been associated with the biological activity. Optimization of the activity and specificity of the AMPs using large peptide libraries is a tedious and expensive route. In this venture, QSAR can be used to shed light or reveal the structural features that should be incorporated in the design of new AMPs. However within the realm of QSAR, 3D-QSAR of peptides is an overwhelming task due to the sheer number of conformational degrees of freedom for peptides. To achieve this, we propose the use of a validated 2D-QSAR technique coined HomoSAR that is specifically designed for peptide QSAR. It has the ability to extract all necessary information from a set of peptides to elucidate the underlying structure activity relationships, based on homology principles and similarity techniques. The present work is a comprehensive study on a dataset of protegrin antimicrobial peptides isolated from porcine leukocytes with a broad spectrum of activity against both Gram-positive and Gram-negative bacteria, as well as the fungus C. albicans and HIV-1 virus. The HomoSAR models for antimicrobial activity against six different species highlighted two major determinants of activity; firstly the optimal length of protegrins for exhibiting broad-spectrum antimicrobial activity against bacteria and fungi is 16 residues. Secondly, for antimicrobial activity against the yeast C. albicans, it turns out that it is the electronic property that should be tempered to modulate activity. This is not a major attribute for both Gram negative and Gram positive bacteria.

 Please wait while we load your content...
Please wait while we load your content...