Integration of pathway structure information into a reweighted partial Cox regression approach for survival analysis on high-dimensional gene expression data†
Abstract
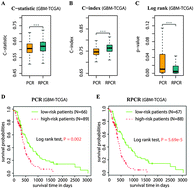
Accurately predicting the risk of cancer relapse or death is important for clinical utility. The emerging high-dimensional gene expression data provide the opportunity as well as the challenge to uncover the relationship between gene expression and censored survival outcome. While several Cox models have been proposed to deal with high-dimensional covariates and censored continuous survival data, they usually generalize poorly to independent datasets. Most methods build the Cox model exclusively on gene expression data, but ignore the molecular interaction relation among genes, which has been successfully integrated into molecular classification with categorical outcomes and improved predictive performance. Here, we integrate gene-interaction information into a Cox model and propose a reweighted partial Cox regression (RPCR) approach in order to accurately predict the risk of cancer events. RPCR improves the predictive accuracy and generalization of a Cox model by promoting genes with large topological importance, which is evaluated by a directed random walk in a reconstructed global pathway graph. We applied RPCR to the survival prediction of two cancer types and used two concordance statistic measures to assess the prediction performance. Both within-dataset experiments and cross-dataset experiments showed that RPCR could predict the risk of patients with higher accuracy and greater robustness.

 Please wait while we load your content...
Please wait while we load your content...