A Gaussian network model study suggests that structural fluctuations are higher for inactive states than active states of protein kinases†
Abstract
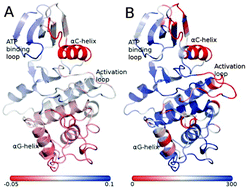
We performed Gaussian network model based normal mode analysis of 3-dimensional structures of multiple active and inactive forms of protein kinases. In 14 different kinases, a more number of residues (1095) show higher structural fluctuations in inactive states than those in active states (525), suggesting that, in general, mobility of inactive states is higher than active states. This statistically significant difference is consistent with higher crystallographic B-factors and conformational energies for inactive than active states, suggesting lower stability of inactive forms. Only a small number of inactive conformations with the DFG motif in the “in” state were found to have fluctuation magnitudes comparable to the active conformation. Therefore our study reports for the first time, intrinsic higher structural fluctuation for almost all inactive conformations compared to the active forms. Regions with higher fluctuations in the inactive states are often localized to the αC-helix, αG-helix and activation loop which are involved in the regulation and/or in structural transitions between active and inactive states. Further analysis of 476 kinase structures involved in interactions with another domain/protein showed that many of the regions with higher inactive-state fluctuation correspond to contact interfaces. We also performed extensive GNM analysis of (i) insulin receptor kinase bound to another protein and (ii) holo and apo forms of active and inactive conformations followed by multi-factor analysis of variance. We conclude that binding of small molecules or other domains/proteins reduce the extent of fluctuation irrespective of active or inactive forms. Finally, we show that the perceived fluctuations serve as a useful input to predict the functional state of a kinase.

 Please wait while we load your content...
Please wait while we load your content...