lncRNA-MFDL: identification of human long non-coding RNAs by fusing multiple features and using deep learning†
Abstract
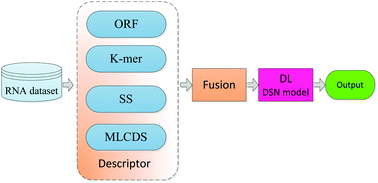
Long noncoding RNAs (lncRNAs) are emerging as a novel class of noncoding RNAs and potent gene regulators, which play an important and varied role in cellular functions. lncRNAs are closely related with the occurrence and development of some diseases. High-throughput RNA-sequencing techniques combined with de novo assembly have identified a large number of novel transcripts. The discovery of large and ‘hidden’ transcriptomes urgently requires the development of effective computational methods that can rapidly distinguish between coding and long noncoding RNAs. In this study, we developed a powerful predictor (named as lncRNA-MFDL) to identify lncRNAs by fusing multiple features of the open reading frame, k-mer, the secondary structure and the most-like coding domain sequence and using deep learning classification algorithms. Using the same human training dataset and a 10-fold cross validation test, lncRNA-MFDL can achieve 97.1% prediction accuracy which is 5.7, 3.7, and 3.4% higher than that of CPC, CNCI and lncRNA-FMFSVM predictors, respectively. Compared with CPC and CNCI predictors in other species (e.g., anole lizard, zebrafish, chicken, gorilla, macaque, mouse, lamprey, orangutan, xenopus and C. elegans) testing datasets, the new lncRNA-MFDL predictor is also much more effective and robust. These results show that lncRNA-MFDL is a powerful tool for identifying lncRNAs. The lncRNA-MFDL software package is freely available at http://compgenomics.utsa.edu/lncRNA_MDFL/ for academic users.

 Please wait while we load your content...
Please wait while we load your content...