Affinity proteomics led identification of vimentin as a potential biomarker in colon cancers: insights from serological screening and computational modelling†
Abstract
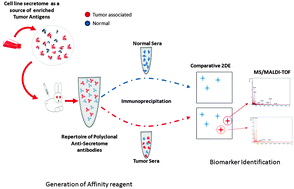
Proteomic analysis using multiplex affinity reagents is perhaps the most reliable strategy to capture differentially expressed proteins that are slightly or immensely modified. In addition to expressional variation, it is comprehensively evident that the immunogenicity of a protein can be a deciding factor for instigating an inflammation afflicted-carcinogenesis. Considering both these factors, a simple and systematic strategy was designed to capture the immunogenic cancer biomarkers from sera of colorectal cancer patients. The affinity reagent, in the form of an antibody repertoire against the secretome of the HT29 cell line was used to grade the sera samples on the basis of the degree of immuno-reactivity and to capture differentially expressed antigens from the patient sera. Following affinity based 2DE-MALDI-TOF; the proteins were identified as (1) soluble vimentin; and (2) TGF-beta-inhibited membrane-associated protein (PP16B), in colon cancer sera and (3) keratin, type II cytoskeletal protein in rectal cancer sera. Pathway reconstruction and protein–protein networking of identified proteins predicted only Vimentin to be physically and genetically engaged in close proximity with the most established colorectal cancer associated tumorigenic pathways. Furthermore, our findings suggest that a possible surface stoichiometric shift in the structure of protein could be due to mutations in the coding sequence of Vimentin that may elicit its enhanced secretion possibly due to protein-hyperphosphorylation. Of the three proteins identified, only Vimentin showed higher expression in sera of colon cancer patients alone. Thus, it could be argued that vimentin might help in predicting individuals at higher risk of developing colon cancers. Our data are therefore suggestive of using vimentin as an antigen for tumor vaccination in an autologous set-up for colon cancers.

 Please wait while we load your content...
Please wait while we load your content...