Unraveling the complexity of protein backbone dynamics with combined 13C and 15N solid-state NMR relaxation measurements†
Abstract
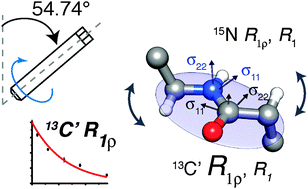
Typically, protein dynamics involve a complex hierarchy of motions occurring on different time scales between conformations separated by a range of different energy barriers. NMR relaxation can in principle provide a site-specific picture of both the time scales and amplitudes of these motions, but independent relaxation rates sensitive to fluctuations in different time scale ranges are required to obtain a faithful representation of the underlying dynamic complexity. This is especially pertinent for relaxation measurements in the solid state, which report on dynamics in a broader window of time scales by more than 3 orders of magnitudes compared to solution NMR relaxation. To aid in unraveling the intricacies of biomolecular dynamics we introduce 13C spin–lattice relaxation in the rotating frame (R1ρ) as a probe of backbone nanosecond-microsecond motions in proteins in the solid state. We present measurements of 13C′ R1ρ rates in fully protonated crystalline protein GB1 at 600 and 850 MHz 1H Larmor frequencies and compare them to 13C′ R1, 15N R1 and R1ρ measured under the same conditions. The addition of carbon relaxation data to the model free analysis of nitrogen relaxation data leads to greatly improved characterization of time scales of protein backbone motions, minimizing the occurrence of fitting artifacts that may be present when 15N data is used alone. We also discuss how internal motions characterized by different time scales contribute to 15N and 13C relaxation rates in the solid state and solution state, leading to fundamental differences between them, as well as phenomena such as underestimation of picosecond-range motions in the solid state and nanosecond-range motions in solution.
- This article is part of the themed collection: Celebrating the 2016 RSC Prize and Award Winners


 Please wait while we load your content...
Please wait while we load your content...