Block ionomer complexes consisting of siRNA and aRAFT-synthesized hydrophilic-block-cationic copolymers: the influence of cationic block length on gene suppression†‡
Abstract
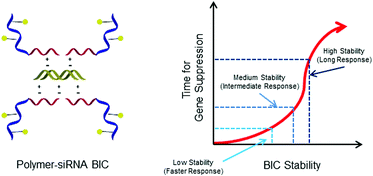
Block ionomer complex (BIC) dissociation and the subsequent effects on gene “knockdown” are reported. Aqueous reversible addition–fragmentation chain transfer (aRAFT) polymerization was utilized to prepare a statistical macro chain transfer agent (macroCTA) consisting of N-(2-hydroxypropyl)methacrylamide (HPMA) and N-(3-aminopropyl)methacrylamide (APMA); HPMA confers water stability, and APMA provides a facile pathway for the conjugation of folic acid, a cellular targeting moiety. This macroCTA was chain extended with N,N-(3-dimethylaminopropyl)methacrylamide (DMAPMA), thus preparing hydrophilic-block-cationic copolymers with varying repeat units of DMAPMA. After end-group removal and folic acid conjugation, these hydrophilic-block-cationic copolymers were complexed with small interfering RNA (siRNA) and GLuc DNA, a double stranded DNA (dsDNA) analogue of siRNA. These complexes were prepared at a nitrogen-to-phosphate ratio (N : P) = 1, and complexes prepared with siRNA and GLuc DNA were demonstrated to be comparable; the hydrodynamic radii (Rh) and changes to the secondary structure were identical, while ζ-potential and gel electrophoresis confirmed complex neutrality. Analytical ultracentrifugation (AUC) was utilized to ascertain binding constants and stoichiometry. Increased DMAPMA block length (cationic length) caused an increase in the binding constant, but the stoichiometry remained constant at 1 : 1. Solution differential scanning calorimetry (DSC) was conducted to investigate the BIC stability. Similar to AUC, the melting temperature (Tm) increased with increasing cationic block length, and overall, a shift in Tm of ∼40 °C was observed, indicating that increasing the DMAPMA length confers greater BIC stability. Furthermore, complex dissociation was not observed. Gene down-regulation was monitored in KB cells expressing Gaussia Luciferase, and the time for maximum gene knockdown to occur increased with increasing DMAPMA block length. Given the large binding constants and this increased stability, it can be concluded that in vitro complex dissociation occurs via an ion exchange mechanism.

 Please wait while we load your content...
Please wait while we load your content...