Predicting protein–protein interactions between human and hepatitis C virus via an ensemble learning method†
Abstract
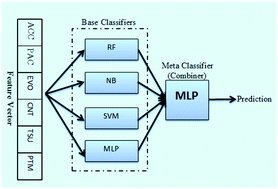
An estimated 170 million people, approximately 3% of the world population, are chronically infected with the hepatitis C virus (HCV). More than 350 000 deaths are reported annually, which are caused by HCV. HCV, similar to a variety of viruses, causes disease in humans by altering protein–protein interactions within the host cells. Experimental approaches for the detection of host–virus PPIs have many inherent limitations. Computational approaches to predict these interactions are therefore of significant importance. While many studies have been developed to predict intra-species PPIs in the last decade, predictions on inter-species PPIs such as human–HCV PPIs are rare. In this study, we developed an ensemble learning method to predict PPIs between human and HCV proteins. Our model utilises four well-established diverse learners as base classifiers including random forest (RF), Naïve Bayes (NB), support vector machine (SVM) and multilayer perceptron (MLP). In addition, an MLP was used as a meta-learner to combine base learners' predictions to provide the final prediction. To encode human and HCV proteins as feature vectors, we used six different descriptors as follows: amino acid composition (ACC), pseudo amino acid composition (PAC), evolutionary information feature, network centrality measures, tissue information and post-translational modification information. To assess the prediction power of the proposed method, we assembled a benchmark dataset composed of confident positive and negative PPIs. In a 10-fold cross-validation experiment, our prediction method achieved accuracy and specificity as high as 83% and 94%, respectively. Furthermore, in an independent test set the proposed method achieved an accuracy of 84% and a specificity of 92%. When compared with the existing method, our method showed a better performance. These results revealed that our method is suitable for performing PPI prediction in a host–pathogen context.

 Please wait while we load your content...
Please wait while we load your content...