Ensemble learning prediction of protein–protein interactions using proteins functional annotations†
Abstract
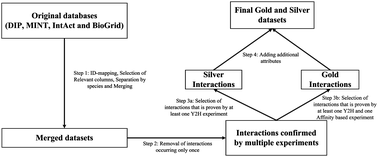
Protein–protein interactions are important for the majority of biological processes. A significant number of computational methods have been developed to predict protein–protein interactions using protein sequence, structural and genomic data. Vast experimental data is publicly available on the Internet, but it is scattered across numerous databases. This fact motivated us to create and evaluate new high-throughput datasets of interacting proteins. We extracted interaction data from DIP, MINT, BioGRID and IntAct databases. Then we constructed descriptive features for machine learning purposes based on data from Gene Ontology and DOMINE. Thereafter, four well-established machine learning methods: Support Vector Machine, Random Forest, Decision Tree and Naïve Bayes, were used on these datasets to build an Ensemble Learning method based on majority voting. In cross-validation experiment, sensitivity exceeded 80% and classification/prediction accuracy reached 90% for the Ensemble Learning method. We extended the experiment to a bigger and more realistic dataset maintaining sensitivity over 70%. These results confirmed that our datasets are suitable for performing PPI prediction and Ensemble Learning method is well suited for this task. Both the processed PPI datasets and the software are available at http://sysbio.icm.edu.pl/indra/EL-PPI/home.html.

 Please wait while we load your content...
Please wait while we load your content...