A sensitive microfluidic platform for a high throughput DNA methylation assay
Abstract
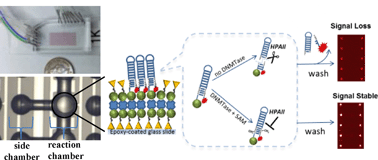
DNA methylation is an epigenetic modification essential for normal development and maintenance of somatic biological functions. DNA methylation provides heritable, long-term chromatin regulation and the aberrant methylation pattern is associated with complex diseases including cancer. Discovering novel therapeutic targets demands development of high-throughput, sensitive and inexpensive screening platforms for libraries of chemical or biological matter involved in DNA methylation establishment and maintenance. Here, we present a universal, high-throughput, microfluidic-based fluorometric assay for studying DNA methylation in vitro. The enzymatic activity of bacterial HPAII DNA methyltransferase and its kinetic properties are measured using the assay (KDNAm = 5.8 nM, KSAMm = 9.8 nM and Kcat = 0.04 s−1). Using the same platform, we then demonstrate a two-step approach for high-throughput in vitro identification and characterization of small molecule inhibitors of methylation. The approach is examined using known non-nucleoside inhibitors, SGI-1027 and RG108, for which we measured IC50 of 4.5 μM and 87.5 nM, respectively. The dual role of the microfluidic-based methylation assay both for the quantitative characterization of enzymatic activity and high-throughput screening of non-nucleoside inhibitors coupled with quantitative characterization of the inhibition potential highlights the advantages of our system for epigenetic studies.

 Please wait while we load your content...
Please wait while we load your content...