M1.3 – a small scaffold for DNA origami †
Abstract
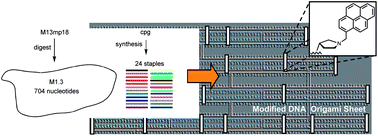
The DNA origami method produces programmable nanoscale objects that form when one long scaffold strand hybridizes to numerous oligonucleotide staple strands. One scaffold strand is dominating the field: M13mp18, a bacteriophage-derived vector 7249 nucleotides in length. The full-length M13 is typically folded by using over 200 staple oligonucleotides. Here we report the convenient preparation of a 704 nt fragment dubbed “M1.3” as a linear or cyclic scaffold and the assembly of small origami structures with just 15–24 staple strands. A typical M1.3 origami is large enough to be visualized by TEM, but small enough to show a cooperativity in its assembly and thermal denaturation that is reminiscent of oligonucleotide duplexes. Due to its medium size, M1.3 origami with globally modified staples is affordable. As a proof of principle, two origami structures with globally 5′-capped staples were prepared and were shown to give higher UV-melting points than the corresponding assembly with unmodified DNA. M1.3 has the size of a gene, not a genome, and may function as a model for gene-based nanostructures. Small origami with M1.3 as a scaffold may serve as a workbench for chemical, physical, and biological experiments.

 Please wait while we load your content...
Please wait while we load your content...