A design automation framework for computational bioenergetics in biological networks†
Abstract
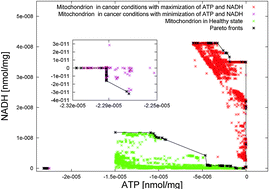
The bioenergetic activity of mitochondria can be thoroughly investigated by using computational methods. In particular, in our work we focus on ATP and NADH, namely the metabolites representing the production of energy in the cell. We develop a computational framework to perform an exhaustive investigation at the level of species, reactions, genes and metabolic pathways. The framework integrates several methods implementing the state-of-the-art algorithms for many-objective optimization, sensitivity, and identifiability analysis applied to biological systems. We use this computational framework to analyze three case studies related to the human mitochondria and the algal metabolism of Chlamydomonas reinhardtii, formally described with algebraic differential equations or flux balance analysis. Integrating the results of our framework applied to interacting organelles would provide a general-purpose method for assessing the production of energy in a biological network.
- This article is part of the themed collection: Molecular BioSystems Network Biology

 Please wait while we load your content...
Please wait while we load your content...