Broadening the signal specificity of prokaryotic promoters by modifying cis-regulatory elements associated with a single transcription factor
Abstract
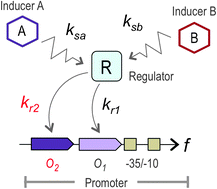
In this report, we experimentally demonstrate that improving the cis-regulatory region of a target promoter can significantly enhance the response to an otherwise poor inducer. The transcriptional factor (TF) BenR of Pseudomonas putida KT2440 is a member of the AraC/XylS family that activates the Pb promoter in response to benzoate. This TF can also trigger Pb activity in response to 3-methylbenzoate (3MBz), but with lower efficiency. Unlike other family members, BenR appears to recognize one operator partially overlapping the −35 promoter region and which is followed by an upstream DNA sequence that lacks an essential motif for TF binding. By generating a promoter variant composed of two complete operator sequences, we observed an enhancement in the sensitivity of Pb to the two inducers. This effect was more pronounced in the case of 3MBz, for which the transcriptional response was approximately 4–5 times higher with the variant than with the wild type promoter. By comparing the responses of the promoters to different concentrations of the inducers, we observed that the modification of the BenR binding region changes the inherent logic of Pb from an amplifier-like behaviour, in which benzoate acts as the sole input, towards OR-gate behaviour, in which 3MBz acts as a second input. Using a mathematical model, we deduced that the second binding site engineered in the Pb promoter enhances the activity of BenR that is bound to the natural operator region, increasing the inducing sensitivity. This work demonstrates how the promiscuity or specificity of inducer recognition can be tuned in a regulatory network without TF mutation and suggests new strategies for the engineering of logic operations in living systems.

 Please wait while we load your content...
Please wait while we load your content...