Generating multiplex gradients of biomolecules for controlling cellular adhesion in parallel microfluidic channels†
Abstract
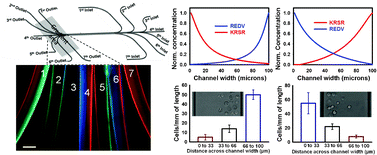
Here we present a microfluidic platform to generate multiplex gradients of biomolecules within parallel microfluidic channels, in which a range of multiplex concentration gradients with different profile shapes are simultaneously produced. Nonlinear polynomial gradients were also generated using this device. The gradient generation principle is based on implementing parrallel channels with each providing a different hydrodynamic resistance. The generated biomolecule gradients were then covalently functionalized onto the microchannel surfaces. Surface gradients along the channel width were a result of covalent attachments of biomolecules to the surface, which remained functional under high shear stresses (50 dyn/cm2). An IgG antibody conjugated to three different fluorescence dyes (FITC, Cy5 and Cy3) was used to demonstrate the resulting multiplex concentration gradients of biomolecules. The device enabled generation of gradients with up to three different biomolecules in each channel with varying concentration profiles. We were also able to produce 2-dimensional gradients in which biomolecules were distributed along the length and width of the channel. To demonstrate the applicability of the developed design, three different multiplex concentration gradients of REDV and KRSR peptides were patterned along the width of three parallel channels and adhesion of primary human umbilical vein endothelial cell (HUVEC) in each channel was subsequently investigated using a single chip.

 Please wait while we load your content...
Please wait while we load your content...