Proteochemometric modeling as a tool to design selective compounds and for extrapolating to novel targets
Abstract
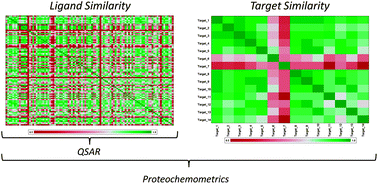
‘Proteochemometric modeling’ is a bioactivity modeling technique founded on the description of both small molecules (the ligands), and
* Corresponding authors
a Division of Medicinal Chemistry, Leiden/Amsterdam Center for Drug Research, Einsteinweg 55, Leiden, The Netherlands
b Tibotec BVBA, Turnhoutseweg 30, Belgium
c
Unilever Centre for Molecular Science Informatics, Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge, United Kingdom
E-mail:
ab454@cam.ac.uk
Tel: +44 (1223) 762 983
‘Proteochemometric modeling’ is a bioactivity modeling technique founded on the description of both small molecules (the ligands), and

 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
G. J. P. van Westen, J. K. Wegner, A. P. IJzerman, H. W. T. van Vlijmen and A. Bender, Med. Chem. Commun., 2011, 2, 16 DOI: 10.1039/C0MD00165A
To request permission to reproduce material from this article, please go to the Copyright Clearance Center request page.
If you are an author contributing to an RSC publication, you do not need to request permission provided correct acknowledgement is given.
If you are the author of this article, you do not need to request permission to reproduce figures and diagrams provided correct acknowledgement is given. If you want to reproduce the whole article in a third-party publication (excluding your thesis/dissertation for which permission is not required) please go to the Copyright Clearance Center request page.
Read more about how to correctly acknowledge RSC content.
 Fetching data from CrossRef.
Fetching data from CrossRef.
This may take some time to load.
Loading related content
