Quantitative and sensitive detection of rare mutations using droplet-based microfluidics†
Abstract
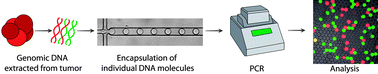
Somatic mutations within tumoral DNA can be used as highly specific
* Corresponding authors
a
Institut de Science et d'Ingénierie Supramoléculaires (ISIS), Université de Strasbourg, CNRS UMR 7006, 8 allée Gaspard Monge, BP 70028, Strasbourg Cedex, France
E-mail:
griffiths@unistra.fr, vtaly@unistra.fr
b Université Paris Descartes, INSERM UMR-S775, Centre Universitaire des Saints-Pères, 45 rue des Saints-Pères, Paris Cedex 06, France
c Max-Planck-Institute for Dynamics and Self-organization, Am Fassberg 17, Göttingen, Germany
d RainDance Technologies France, 8 allée Gaspard Monge, BP 70028, Strasbourg Cedex, France
e RainDance Technologies, Lexington, Massachusetts, USA
Somatic mutations within tumoral DNA can be used as highly specific

 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
D. Pekin, Y. Skhiri, J. Baret, D. Le Corre, L. Mazutis, C. Ben Salem, F. Millot, A. El Harrak, J. B. Hutchison, J. W. Larson, D. R. Link, P. Laurent-Puig, A. D. Griffiths and V. Taly, Lab Chip, 2011, 11, 2156 DOI: 10.1039/C1LC20128J
To request permission to reproduce material from this article, please go to the Copyright Clearance Center request page.
If you are an author contributing to an RSC publication, you do not need to request permission provided correct acknowledgement is given.
If you are the author of this article, you do not need to request permission to reproduce figures and diagrams provided correct acknowledgement is given. If you want to reproduce the whole article in a third-party publication (excluding your thesis/dissertation for which permission is not required) please go to the Copyright Clearance Center request page.
Read more about how to correctly acknowledge RSC content.
 Fetching data from CrossRef.
Fetching data from CrossRef.
This may take some time to load.
Loading related content
