Nanochannel confinement: DNA stretch approaching full contour length
Abstract
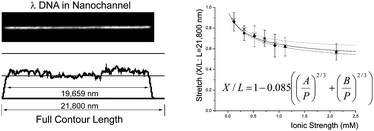
Fully stretched DNA molecules are becoming a fundamental component of new systems for comprehensive genome analysis. Among a number of approaches for elongating DNA molecules, nanofluidic molecular confinement has received enormous attentions from physical and biological communities for the last several years. Here we demonstrate a well-optimized condition that a DNA molecule can stretch almost to its full contour length: the average stretch is 19.1 µm ± 1.1 µm for YOYO-1 stained λ DNA (21.8 µm contour length) in 250 nm × 400 nm channel, which is the longest stretch value ever reported in any nanochannels or nanoslits. In addition, based on Odijk's

 Please wait while we load your content...
Please wait while we load your content...