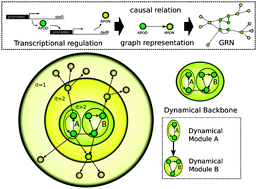
Gene regulatory networks constitute the first layer of the cellular computation for cell adaptation and surveillance. In these webs, a set of causal relations is built up from thousands of interactions between transcription factors and their target genes. The large size of these webs and their entangled nature make it difficult to achieve a global view of their internal organisation. Here, this problem has been addressed through a comparative study of Escherichia coli, Bacillus subtilis and Saccharomyces cerevisiae gene regulatory networks. We extract the minimal core of causal relations, uncovering the hierarchical and modular organisation from a novel dynamical/causal perspective. Our results reveal a marked top-down hierarchy containing several small dynamical modules for E. coli and B. subtilis. Conversely, the yeast network displays a single but large dynamical module in the middle of a bow-tie structure. We found that these dynamical modules capture the relevant wiring among both common and organism-specific biological functions such as transcription initiation, metabolic control, signal transduction, response to stress, sporulation and cell cycle. Functional and topological results suggest that two fundamentally different forms of logic organisation may have evolved in bacteria and yeast.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...