Open Access Article
Open Access ArticleDetection of intracellular microRNA-21 for cancer diagnosis by a nanosystem containing a ZnO@polydopamine and DNAzyme probe†
Yuanyuan Liua,
Ranran Wangb,
Fengxia Zhang a,
Yongshan Ma
a,
Yongshan Ma a and
Tianyi Jiang
a and
Tianyi Jiang *a
*a
aSchool of Municipal and Environmental Engineering, Shandong Jianzhu University, Jinan 250101, Shandong, P. R. China. E-mail: jiangtianyi@sdjzu.edu.cn
bYantai Engineering & Technology College, Yantai 264006, Shandong, P. R. China
First published on 26th April 2024
Abstract
MicroRNAs (miRNAs) are a series of single-stranded non-coding ribonucleic acid (RNA) molecules which associated closely with various human diseases. Efficient strategies for detecting miRNAs are of great significance to cancer diagnosis and prognosis. Here we provide a novel nanosystem that can be applied for the detection of miRNAs. The nanosystem consists of a single-stranded deoxyribonucleic acid (DNA) probe and a probe carrier. The DNA probe was designed based on a deoxyribozyme (DNAzyme) with several necessary functional sequences and two fluorescent dyes labeled at proper sites. The ZnO@polydopamine (ZnO@PDA) nanomaterial serves not only as a probe carrier, but also as a supplier of Zn2+ that can activate the DNAzyme. The DNA probe will undergo a conformation alteration induced by miRNA-21, which then trigger the DNAzyme catalyzed self-cleavage reaction with the assist of Zn2+ provided by ZnO decomposition under weak acid environment. A change of fluorescent color will occur due to the interruption of fluorescence resonance energy transfer between the two fluorescent dyes, and the dissociated miRNA-21 can repeatedly induce the above responses to amplify the fluorescence signal. The feasibility of the whole procedure was demonstrated by various experiments. This nanosystem showed a good selectivity towards miRNA-21, and under the optimal incubation time of 2 hours, a good linear relationship was obtained in a concentration range of 0.01–2.0 nM with a detection limit of 3.8 pM. In in vivo detection, an obvious fluorescence color change from red to green can be observed in the presence of miRNA-21. The results proved that this miRNA detection strategy has a broad application prospect in tumor diagnosis and miRNA related biological studies.
1 Introduction
MicroRNAs (miRNAs) are a series of single-stranded small molecules (approximately 19–23 nucleotides) of non-coding RNA. They serve as crucial regulatory factors in gene expression and play important roles in various cellular processes.1,2 In particular, increasing evidence suggests that dysregulation of miRNA expression is closely associated with various human diseases, such as cancer, and so miRNAs have become biomarkers for cancer diagnosis and prognosis.1,3–5 Therefore, accurate and sensitive detection of miRNAs, especially in situ monitoring of miRNA expression in living cancer cells is of great significance.6–10 However, due to the low abundance of miRNAs in cells and the complex in vivo environment, the development of effective signal amplification strategies is indispensable for intracellular miRNA imaging.11–14Deoxyribozymes (DNAzymes) are a class of single-stranded DNA fragments with catalytic function, which have high catalytic activity and structure recognition ability. The RNA-cleaving DNAzymes are the most well studied DNAzymes. An RNA-cleaving DNAzyme usually contain a catalytic domain and two substrate recognition domains, and the substrate recognition domains contain the necessary nucleotide sequence for specific binding to the substrate. DNAzymes known as 8–17 and 10–23 were originally isolated to cleave the phosphodiester bond of an all-RNA substrate,15,16 while some other DNAzymes, such as 17E, GR-5, 39E and NaA43 were isolated to cleave an adenine ribonucleotide (rA) embedded in an otherwise all-DNA sequence.15,17–20 The activities of DNAzymes generally require metal ions as cofactors since metal ions are important for the creation of stable structures of DNA, and many DNAzymes are specific or highly selective for a given metal ion, such as Pb2+, Mg2+, Cu2+, and Zn2+, etc.21 DNAzymes that are dependent on specific amino acid have also been reported.22 Furthermore, same as the protein enzymes, DNAzymes can also be rationally designed to respond to specific targets.23,24
DNAzymes have broad application prospects in the design of biosensors and nanoprobes owing to the advantages of good stability, low non-specific adsorption, and easy to design, etc.15 They have been studied for direct or indirect sensing of various analytes, including metal ions, toxic gases, nucleic acids, proteins, and bacteria, etc.25–28 Particularly, the unique catalytic mechanism of DNAzymes makes them excellent signal amplifiers for highly sensitive detections of low abundance targets. Over the past few years, quite a few DNAzyme-based amplifying sensing platforms have been built. For example, Wen et al. constructed a molecular machine for DNA amplification detection by combining molecular beacons with DNAzymes catalytic beacons and rolling ring amplification technology.29 As another example, an amplified fluorescence biosensing strategy for serum prostate specific antigen detection was developed based on Zn2+-dependent DNAzyme.30 Such an advantage of DNAzymes-based biosensing techniques meet well with the requirements of miRNAs detection since they are typical low abundance analytes. However, most of these methods are limited to the detection of target miRNAs in the extracellular environment and can rarely be applied in living cells.31–33 In particular, DNAzymes based intracellular sensors have been poorly studied in miRNA imaging. Therefore, it is necessary to develop new signal amplification techniques based on DNAzymes for the detection of low abundance miRNAs in cells.
In order to achieve intracellular detection of miRNAs, methods that can effectively deliver detection probes to the cytoplasm of living cells are also required. Since common nucleic acid probes cannot penetrate cell membranes, various nanomaterials are commonly used to deliver DNA probes through cellular endocytosis.34,35 In this research, a unimolecular probe was designed and synthesized for the in vivo detection of miRNA. The unimolecular consists of a Zn2+-dependent 17E DNAzyme,10 its substrate sequence, and a target-binding sequence, and together these sequences form an intact hairpin structure. The hairpin structure of unimolecular will be opened by the hybridization of target-binding sequence and target miRNA to active the DNAzyme, and then the substrate sequence will be cleaved by DNAzyme to lead to the excitation of fluorescence signal. Furthermore, by using biodegradable ZnO as the core and biocompatible polydopamine (PDA) as the shell, the ZnO@PDA core–shell nanostructures were synthesized as carriers to transport probes into cells. Since ZnO can be degraded into Zn2+ under weak acid condition, the degraded Zn2+ can act as the activator of the DNAzyme, and so there is no need to add additional Zn2+. Based on signal amplification, background interference was effectively reduced, and highly sensitive detection of miRNA-21 (ref. 36) in cells was achieved. This method has broad application prospects in tumor diagnosis and miRNA related biological studies.
2 Experimental
2.1 Chemicals and materials
The DNAzyme containing probe and miRNAs are all synthesized by Sangon Biotech (Shanghai) Co., Ltd. The sequences of the oligonucleotides are listed in as follows:Probe: 5′-![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) AAAAAAACATCAGTATrAGGACTTTTTTTTT
AAAAAAACATCAGTATrAGGACTTTTTTTTT![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) TCTCCGAG-CCGGCTGAAATATGATAAGC-3′
TCTCCGAG-CCGGCTGAAATATGATAAGC-3′
F-DNA: 5′-![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) AAAAAAACATCAGTATrAGGACTTTTTTTTTGTCTCCGAG-CCGGCTGAAATATGATAAGC-3′
AAAAAAACATCAGTATrAGGACTTTTTTTTTGTCTCCGAG-CCGGCTGAAATATGATAAGC-3′
The bold underlined C is modified by a carboxyfluorescein (FAM) group, while the bold underlined G is modified by a carboxy tetramethyl rhodamine (TAMRA) group. The italic rA is the cleavage site of DNAzyme.
miRNA-21: 5′-UAGCUUAUCAGACUGAUGUUGA-3′
miRNA-21b: 5′- UAGUUUAUCAGACUGAUAUUUCC-3′
miRNA-let-7a: 5′-UGAGGUAGUAGGUUGUAUAGUU-3′
miRNA-141: 5′-UAACACUGUCUGGUAAAGAUGG-3′
Nano zinc oxide (ZnO) nanoparticles, dopamine, tris, sodium hydroxide (NaOH) and other chemical reagents are analytically pure and purchased from Sigma-Aldrich (USA). The Dulbecco's modified Eagle's medium (DMEM), RPMI-1640 medium, and fetal bovine serum (FBS) are purchased from Thermo Fisher Scientific Inc. The antibiotics and Gel Red nucleic acid dye are purchased from Beijing Solarbio Science & Technology Co., Ltd. All the water used in the experiment was ultra-pure water.
2.2 Instruments
The morphologies of all samples were recorded by a JEM-2100 high-resolution transmission electron microscope with an accelerating voltage of 200 kV (Hitach, Japan). The hydrodynamic sizes and Zeta potentials were measured by a Zetasizer Nano-ZS analyzer (Malvern Instruments, Malvern, UK). UV-vis absorption spectra were assayed using a UV-5800 UV-vis spectrophotometer (METASH, China). Fluorescence spectra were assayed using an F-7000 spectrometer (Hitach, Japan). Confocal images were taken on an FV1000 confocal laser scanning microscope (Olympus, Japan).2.3 Preparation of ZnO@PDA
10 mg Dopamine was dissolved in 10 mL newly formulated Tris-HCl buffer (pH 8.5) to a concentration of 1 mg mL−1. The commercially available ZnO nanoparticles was prepared into a 1 mg mL−1 ZnO solution, and 1 mL ZnO solution was added to 10 mL dopamine solution (the molar ratio of dopamine to ZnO was approximately 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After continuous stirring at room temperature for 1 h, the PDA shell was formed on the ZnO surface by self-polymerization of dopamine. The prepared ZnO@PDA was centrifuged (15
1). After continuous stirring at room temperature for 1 h, the PDA shell was formed on the ZnO surface by self-polymerization of dopamine. The prepared ZnO@PDA was centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) and washed with equal volume of ultrapure water for 5 times, and then was dried at 100 °C to obtain the core–shell material. The ZnO and ZnO@PDA nanoparticles were imaged by a JEM-2100 high-resolution transmission electron microscope with an accelerating voltage of 200 kV. The hydrodynamic sizes and Zeta potentials of the ZnO and ZnO@PDA nanoparticles were measured by a Zetasizer Nano-ZS analyzer. The UV-vis absorption spectra of ZnO nanoparticles solutions were recorded with a UV-5800 UV-vis spectrophotometer.
000 rpm, 10 min) and washed with equal volume of ultrapure water for 5 times, and then was dried at 100 °C to obtain the core–shell material. The ZnO and ZnO@PDA nanoparticles were imaged by a JEM-2100 high-resolution transmission electron microscope with an accelerating voltage of 200 kV. The hydrodynamic sizes and Zeta potentials of the ZnO and ZnO@PDA nanoparticles were measured by a Zetasizer Nano-ZS analyzer. The UV-vis absorption spectra of ZnO nanoparticles solutions were recorded with a UV-5800 UV-vis spectrophotometer.
2.4 Preparation of probe
The designed functional probe was synthesized by Sangon Biotech (Shanghai) Co., Ltd. Quantitative Tris-HCl buffer (pH 8.5) was added to prepare the probe to a concentration of 10−4 M, and the probe was diluted to the required concentration according to subsequent experiments. The designed probe was annealed by first heating to 95 °C and then cooling naturally to 37 °C to form the desired hairpin structure. The ZnO@PDA nanoparticles solution (0.1 mg mL−1) was mixed with the annealed probe solution (10−4 M) with a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. The mixture was stirred at room temperature for 1 h, and then was centrifuged at 15
10. The mixture was stirred at room temperature for 1 h, and then was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min to remove excess probes and obtain the ZnO@PDA/probe nanosystem.
000 rpm for 10 min to remove excess probes and obtain the ZnO@PDA/probe nanosystem.
2.5 Polyacrylamide gel electrophoresis
Polyacrylamide gel (7.5%) electrophoresis was employed to verify the cleavage reaction in the absence and presence of miRNA-21. Gel stock solution was prepared by dissolving 7.3 g acrylamide and 0.2 g bis(N,N′-methylene diacrylamide) in 25 mL ultrapure water. To prepare the polyacrylamide gel, 4 mL prepared gel stock solution, 8 mL ultrapure water, 4 mL Tris-HCl buffer (1.5 M, pH 8.8), 150 μL (NH4)2S2O8 (10%, w/v), and 15 μL N,N,N′,N′-tetramethylethylenediamine (TEMED) were added successively to the a beaker and mixed. The mixture was poured into a gel casting stand. After the samples were loaded, the gel was electrophoresed for 30 min at 135 V and then stained with Gel Red dye for 1 h away from light. For the cleavage reaction, hp DNA, miRNA-21, and Zn2+ were added to Tris-HCl buffer (pH 8.5) to concentrations of 200 nM, 200 nM, and 500 nM, respectively, if they are needed in specific reaction systems. The reaction systems were incubated at 37 °C for 3 h.2.6 Fluorescence assay
To study the effect of incubation time on in vitro miRNA-21 detection, the probe was dissolved into Tris-HCl buffer at a concentration of 200 nM. After annealing, the probe solution was mixed with miRNA-21 solution with a concentration of 20 nM, and Zn2+ was added to a concentration of 500 nM. The fluorescence spectrum of the mixture was scanned at certain interval times. To study the effect of target concentration on in vitro miRNA-21 detection, different concentrations of miRNA-21 (0 nM, 0.01 nM, 0.05 nM, 0.25 nM, 0.5 nM, 0.8 nM, 1.2 nM, 2.0 nM, 20 nM, 80 nM, and 200 nM) were mixed with 200 nM probe. Zn2+ was added to the system to a concentration of 500 nM and the mixtures were incubated at 37 °C. After 2 hours, fluorescence spectrum assay was performed. The detections of other miRNAs are similar to that of miRNA-21, while the concentrations of miRNAs were uniformly set at 200 nM. The fluorescence was measured with excitation at 490 nm (for the assay of TAMRA alone, the fluorescence was measured with excitation at 545 nm). For the assay of FFAM/FTAMRA, the fluorescence excitation wavelength was 490 nm, and the emission wavelength for FAM or TAMRA was 522 nm or 580 nm, respectively.2.7 Cytotoxicity assay
L-02 cells were inoculated into a 96-well plate and cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS), penicillin (100 μg mL−1) and streptomycin (100 μg mL−1) at 37 °C in a humidified atmosphere containing 5% CO2 for 24 h. Then half of the culture medium was removed and replaced with new medium containing ZnO@PDA/probe (40 μg mL−1) and the cells were incubated for a certain time (0, 6, 12, 18, or 24 h). Subsequently, MTT solution (0.5 mg mL−1) was added to each well and the cells were incubated for another 4 h. After the MTT solution was removed, 150 μL of DMSO was added to each well and the plate was shaken gently for 10 min. The absorbance at 490 nm was measured to evaluate the cell viability.2.8 Live cell miRNA imaging
The MCF-7 and L-02 cells were cultured according to the procedure reported previously.37 MCF-7 cells were cultured in DMEM supplemented with 10% fetal FBS, penicillin (100 μg mL−1) and streptomycin (100 μg mL−1). L-02 cells were cultured in RPMI-1640 with 10% FBS, penicillin (100 μg mL−1) and streptomycin (100 μg mL−1) in a humidified atmosphere containing 5% CO2 at the temperature of 37 °C. For the fluorescence imaging of miRNA-21, 10 μL of the ZnO@PDA/probe nanosystem (0.2 mg mL−1) was added to 90 μL cell cultures and the mixtures were incubated for 2 h at 37 °C. Then, the cells were washed three times with Tris-HCl buffer (pH 8.5) and fluorescence images were obtained using a confocal laser scanning fluorescence microscope.3 Results and discussion
3.1 Probe design and detection strategy
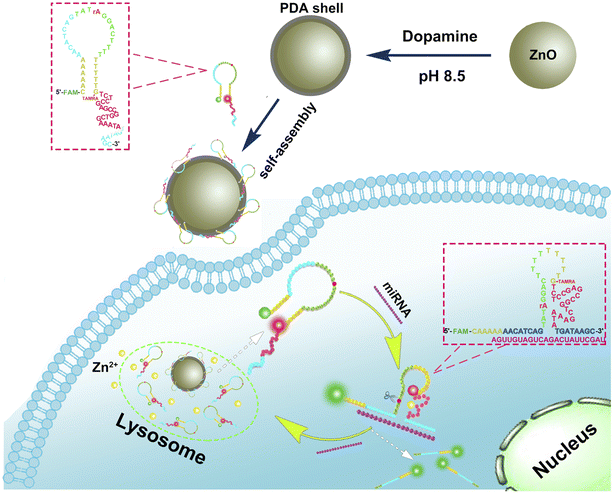
As shown in the schematic (Fig. 1), the single-stranded DNA probe designed in this work consists of the following parts: a DNAzyme sequence (red), a substrate sequence containing a rA cleavage site (green), a complementary sequence of target miRNA (divided in to two parts, blue), and two linker sequences (yellow). All these parts were linked together as a unimolecular probe. The DNAzyme sequence was designed based on the 17E DNAzyme, which has long been applied in biosensing studies.19 Although 17E DNAzyme can utilize several metal ions, such as Pb2+, Zn2+, Mn2+, Co2+, and Mg2+, etc., as a cofactor, only Pb2+ and Zn2+ were proved to induce strong activity of it15,19,38. However, since Pb2+ is generally recognized as highly toxic to cells, Zn2+ was selected as the activator of 17E DNAzyme, which is compatible with the nanomaterial designed in this work. The complementary sequence of target miRNA was designed based on the sequence of miRNA-21, a diagnostic prognostic and therapeutic marker for cancer36,39, while it is possible to be substituted towards other targets. The two linker sequences contain poly T and poly A, respectively, and they could anneal to cause the entire probe to form a hairpin-locked structure. Moreover, at the 5 ′-end of the probe, a cytosine ahead of poly A was modified with green fluorescent dye FAM, and at the middle of the probe, a guanine was modified with red fluorescent dye TAMRA. In the state that the hairpin-locked structure is formed, TAMRA was close to FAM, and thus the fluorescence resonance energy transfer (FRET) process occurred, leading to the decrease of green fluorescence and increase of red fluorescence.As a carrier that transport probes into cells, ZnO@PDA core–shell structure material was synthesized by using dopamine self-polymerization on ZnO surface at pH 8.5. Due to π–π stacking and hydrogen bonding, the probes could be adsorbed on the surface of ZnO@PDA and then be transported into the cell through cellular endocytosis. After entering the cell, the ZnO cores will be decomposed into Zn2+ under the action of intracellular lysosomes, leading to the dissociation of probes and the delivery of probes into the cytoplasm. When the target miRNA (purple) is present, it will combine with its complementary sequence in the probe and open the hairpin to form the active secondary structure of the DNAzyme. Meanwhile, the change of secondary structure will lead to the exposition of cleavage site (rA) in the substrate sequence to the catalytic core of DNAzyme, and then the DNAzyme activated by Zn2+ will cleave the self-strand to divide the probe into two fragments. After the cleavage, the green fluorophores FAM carried by the shorter fragment moves far away from the red fluorophores TAMRA carried by the longer fragment, and the interruption of FRET channel will cause the green signal to recover and the red signal to attenuate. Since the complementary sequence of target miRNA is also separated by the cleavage, its affinity to target miRNA will reduce, and the released miRNA continues to combine with other probes to undergo the above reaction. The reaction cycle enhances the green signal and weakens the red signal continuously, which could achieve the ratio fluorescence monitoring of miRNA in cells. Compared with traditional methods for miRNA detection, this technique that combined the DNAzyme assisted circulatory signal amplification with ZnO@PDA core–shell structure is not only conducive to the transmission of probes into cells, but also independent of additional cofactor to activate DNAzymes. Furthermore, ratiometric fluorescence technology is used to effectively reduce the interference of background signals.
3.2 Synthesis and characterization of ZnO@PDA
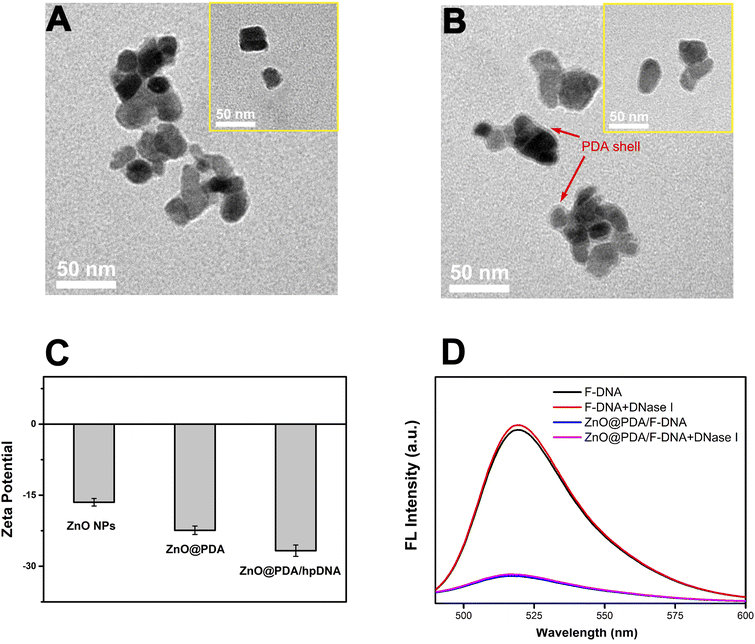
To deliver the probes into the live cells efficiently, a suitable carrier material is needed. In this study, ZnO@PDA core–shell nanomaterial was selected as probe transport carrier. The ZnO nanoparticles has the advantages of low cost and low toxicity to cells, and more importantly, it coordinates well with the Zn2+-dependent DNAzyme used in the probe since it can be completely decomposed to provide Zn2+ in aqueous solution under acidic conditions.40 The cellular endocytosis will deliver the ZnO nanoparticles into lysosome, which has an acid microenviroment (pH 4.5–5.0) and can trigger the decomposition of ZnO.41 The PDA shell of the nanomaterial provides an outer layer for the adsorption of DNA probes owing to its abundant catechol and amino groups. Furthermore, its biocompatibility will further facilitate cellular endocytosis of the material.42The synthesized ZnO@PDA was firstly characterized by transmission electron microscopy (TEM). Compared with that of ZnO nanoparticles (ZnO NPs) (Fig. 2A), an outer layer with lower grayscale surrounding the particles can be seen on the TEM image of ZnO@PDA (Fig. 2B), indicating that the ZnO NPs has been coated by the PDA shell. Moreover, the dynamic light scattering (DLS) results showed that the dispersed ZnO NPs has a size smaller than that of PDA coated ZnO. The hydration radiuses of ZnO NPs and ZnO@PDA were estimated to be 43.8 ± 3.2 nm and 54.7 ± 3.7 nm, respectively (Fig. S1†), which also prove that the ZnO core has been successfully coated by the PDA shell. It is notable that this size is within the range that can be taken up by cells, and so ZnO@PDA can be used as a carrier to transport probes into cells.
The self-polymerization of dopamine and absorbance of DNA probe on the surface of nanomaterial could cause the decrease of Zeta potential.43 So, we also used Zeta potential to characterize ZnO@PDA carrying probe DNA. As shown in Fig. 2C, the Zeta potential of ZnO NPs was −16.8 ± 0.8 mV, which became more negative after dopamine was self-polymerized on ZnO surface (−22.4 ± 0.9 mV), and this change may be due to the hydroxyl groups exposed on the surface of PDA. When the hairpin-locked probe DNA (hp DNA), as negatively charged molecules, were adhered to the ZnO@PDA surface by π–π stacking, the Zeta potential decreased further to −26.7 ± 1.2 mV. These results validate the successful construction of ZnO@PDA nanomaterial and its adsorption of DNA probes.
Meanwhile, the ability of ZnO@PDA nanomaterial to protect DNA probes against nuclease was tested. A single-stranded DNA similar to the probe but was only labeled with FAM at the 5′-end (F-DNA) was synthesized, and the F-DNA in free state or adsorbed by ZnO@PDA was incubated with DNase I, respectively. It can be seen in Fig. 2D that F-DNA exhibited a fluorescence peak at about 520 nm, and the DNase I digested free F-DNA exhibited a similar fluorescence spectrum. When F-DNA was adsorbed by ZnO@PDA, its fluorescence was quenched by the nanomaterial. The incubation of ZnO@PDA/F-DNA complex with DNase I did not cause the recovery of fluorescence. The results prove that ZnO@PDA can efficiently protect the DNA probe from being digested by nuclease. This protection effect might be attributed to the tight combination of the probe and the ZnO@PDA nanomaterial, which produce steric hindrance on the surface against DNase I. The ability of ZnO@PDA to protect DNA probes will facilitate the system to be applied in complex intracellular environment.
3.3 Feasibility of ratiometric fluorescence assay for miRNAs
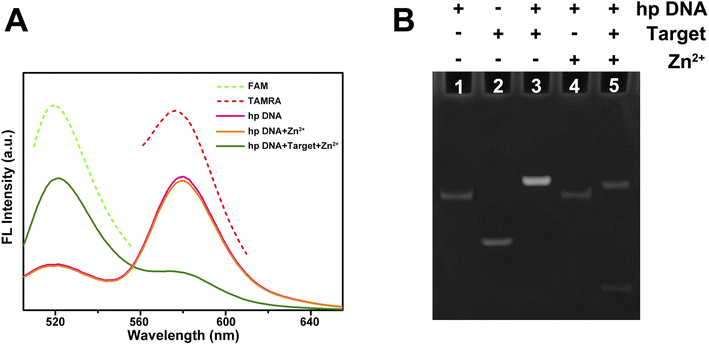
In order to verify the feasibility of utilizing the probe to achieve ratiometric fluorescence assay for miRNAs, several experiments have been conducted. First, fluorescence assay was applied to verify that the fluorescence of probe will change as expected. Two fluorescent dyes with distinct colors, FAM (green) and TAMRA (red), have been labeled at different sites of the probe sequence. As shown in Fig. 3A, the individual FAM (green dotted line) exhibited a fluorescence emission peak at about 520 nm, while the individual TAMRA (red dotted line) exhibited a fluorescence emission peak at about 580 nm. The formation of hairpin-locked structure of probe brought the two fluorescent dyes close to each other and led to the occurrence of FRET process. Consistently, the fluorescence emission spectrum of hp DNA (amaranth line) was generally formed by the superposition of FAM and TAMRA spectra, while the TAMRA emission peak was obvious and the FAM emission peak was rather low. When the hp DNA, target miRNA and Zn2+ were incubated together, the activity of DNAzyme was induced by the target with the assist of Zn2+. The self-cleavage reaction led to the dissociation of FAM labeled fragment from the probe, and the interruption of FRET process resulted in an increased FAM emission at 520 nm accompanied with a decreased TAMRA emission at 580 nm (green line). As a control, the hp DNA incubated with only Zn2+ (orange line) exhibited no significantly change compared to that of individual hp DNA, indicating that such a change of fluorescence emission spectrum was inseparable from the action of target miRNA.Next, the gel electrophoresis was also performed to verify the self-cleavage reaction of probe DNA triggered by target miRNA and Zn2+. As shown in Fig. 3B, the bands in lane 1 and lane 2 with different sizes correspond to the individual hp DNA or target miRNA, respectively. When they were mixed, a band with a size larger than either of them was detected (lane 3), indicating the combination of probe DNA and target miRNA. When only hp DNA and Zn2+ were added to the system (lane 4), the conformation of hp DNA did not change and presented a band similar to lane 1. When all of hp DNA, Zn2+ and target miRNA were mixed together (lane 5), two bands were detected, corresponding to the two products of self-cleavage reaction of the probe. This result further validates the feasibility of this strategy for miRNA detection.
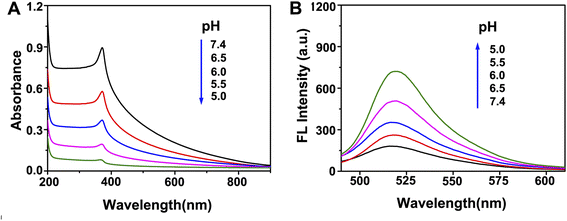
The ZnO@PDA core–shell nanomaterial was used as a carrier to carry the probes into cells in this research. The nanomaterial will be transported into lysosome that has a weak acidic environment, and the ZnO core is expected to be degraded to Zn2+ under this condition to provide cofactors for the activation of DNAzyme. In order to validate this expectation, ZnO was added to the buffer of different pH (5.0, 5.5, 6.0, 6.5, 7.4) and the absorbances of solutions were recorded. As shown in Fig. 4A, after being placed under room temperature for 60 min, the UV absorption of ZnO at about 370 nm exhibited significant differences with changes in pH, proving that the decomposition of ZnO was enhanced with the increase of acidity. The UV absorbance changes at 370 nm of these solutions as a function of time were also recorded. It can be seen in Fig. S2† that ZnO remained stable at pH 7.4 for 120 min, while more than 80% of ZnO was decomposed into Zn2+ after 60 min at pH 5.0.
Fluorescence assay was also applied to validate the decomposition of ZnO. The fluorescence of F-DNA was quenched due to the FRET to PDA when F-DNA was adsorbed on the surface of ZnO@PDA. When the ZnO@PDA/F-DNA system was added to buffer with different pH, the fluorescence of F-DNA recovered in acidic environment, and the recovering of fluorescence became stronger with the gradual decrease of pH (Fig. 4B). This result indicates that the ZnO@PDA nanostructure collapsed and the F-DNA molecules were released in acidic environment due to the decomposition of ZnO. According to the change trend of fluorescence intensity, most fluorescence recovered after 60 min (Fig. S3†), which is consistent with the results of ZnO UV absorption assay. These results demonstrate the feasibility of ZnO core as a supplier of Zn2+ to activate DNAzyme in the cells.
3.4 Detection of miRNA-21 in vitro
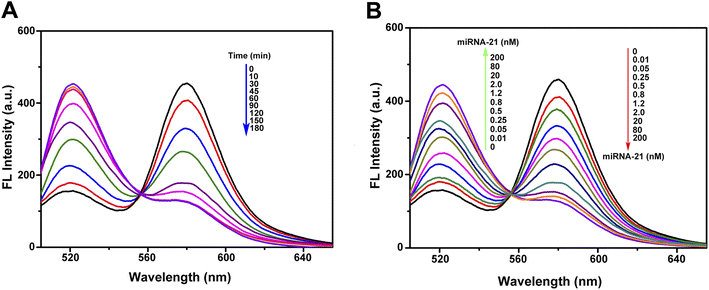
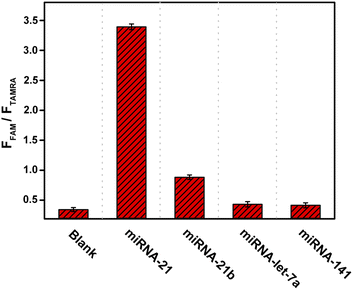
The probe designed in this work is specific to miRNA-21. This microRNA has been demonstrated to have a potential oncogenic function and target tumor inhibitor proteins in almost all types of cancer, and it has been studied in terms of cell proliferation, migration, invasion, metastasis, and apoptosis regulation.39 The expanding role of miRNA-21 makes the development of its detection technology important. Nevertheless, the specificity of probe is determined by the complementary sequence of target (Fig. 1), and so the detection target can be easily changed by sequence replacement. To achieve efficient detection of the target, the effects of incubation time and target concentration on detection were optimized. Fig. 5A shows that with the increase of incubation time, the red fluorescence decreased while the green fluorescence increased continuously due to the cyclic self-cleavage reaction, and the fluorescence recovery basically reached saturation after incubation for 2 hours (Fig. S4†). Fig. 5B shows that under the same incubation time (2 h), the increase of miRNA-21 concentration also led to the decrease of red fluorescence and increase of green fluorescence. The detection results of miRNA-21 showed a good linear relationship (R2 = 0.9946) in a concentration range of 0.01–2.0 nM, and the detection limit was estimated to be 3.8 pM (Fig. S5†).On the other hand, the ability of probe to distinguish different miRNAs is critical to its performance in practical applications. To verify the selectivity of the probe to miRNA-21, we selected miRNA-21b44, which has relative high sequence similarity to miRNA-21, and two completely different miRNAs (miRNA-let-7a45 and miRNA-141 (ref. 46)) as controls. Although there are five base differences between miRNA-21 and miRNA-21b, only two of them are in the regions that are complementary to probe sequence. Fig. 6 shows that as judged by the ratio of FAM to TAMRA fluorescence intensity, the technique has good selectivity towards miRNA-21, and even two base differences with miRNA-21 can be identified, which confirms the ability of this technique to detect miRNA-21 in complex cellular environments.
3.5 Imaging of intracellular miRNA-21
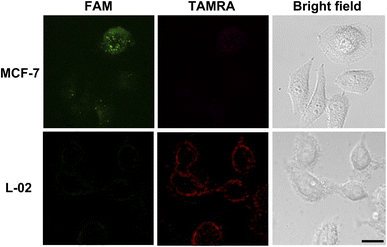
After demonstrating the ability of the probe to detect miRNA-21 in vitro, the ZnO@PDA/probe system was investigated for intracellular miRNA-21 imaging. First, an MTT assay was used for testing the toxicity of the ZnO@PDA/probe nanosystem towards L-02 cell (normal human hepatocyte cell). The result shows that more than 90% of the cells remained viable after 24 hours of incubation with ZnO@PDA/probe, thus proving that the nanosystem could be applied to intracellular fluorescence imaging (Fig. S6†). The human breast adenocarcinoma cell (MCF-7) and L-02 cell with different miRNA-21 expression levels were selected for the intracellular miRNA-21 assays. miRNA-21 has been reported to over-express in MCF-7 cells but has a low expression level in L-02 cells.37 After being incubated with the ZnO@PDA/probe for 2 h, the cell cultures of MCF-7 and L-02 were imaged by laser confocal microscope. As shown in Fig. 7, the green fluorescence signal was obivious while the red fluorescence signal was basically undetectable for MCF-7 cells, indicating the ZnO@PDA/probe system was endocytosed into cells and the Zn2+-dependent self-cleavage reaction of released probes was successfully activated by the combination of intracellular miRNA-21. In contrast, only red fluorescence signal could be observed for L-02 cells. Although the ZnO@PDA/probe system was also endocytosed into L-02 cells, the released probes kept their hairpin-locked structure in the absence of intracellular miRNA-21. These results were consistent with those previously reported for miRNA-21 expression levels in MCF-7 and L-02 cells, and moreover, they demonstrated that this method can monitor the expression of miRNA-21 in cells, which is of great significance for the diagnosis and treatment of cancer.4 Conclusion
In this work, a ZnO@PDA/probe nanosystem used for miRNA-21 detection was designed and characterized. A single-stranded DNA containing a Zn2+-dependent DNAzyme, its substrate sequence, and a target-binding sequence was used as the detection probe. The ZnO@PDA core–shell nanomaterial acts as a carrier and protector of probes. The successful construction of ZnO@PDA was verified by TEM imaging and DLS analysis. Zeta potential analysis proved the adsorption of DNA probe by ZnO@PDA, and it can effectively protect DNA probe from being digested by nuclease. Under weak acidic environment similar to that in lysosome, the ZnO was proved to decompose to provide Zn2+. The results of fluorescent scanning and gel electrophoresis showed that the self-cleavage reaction of probe was triggered by miRNA-21 with the assist of Zn2+, and as expected, the fluorescent color of probe changed from red to green. Furthermore, the change in fluorescence color became more obvious over time, indicating that the reaction can be triggered repeatedly to achieve ratiometric fluorescence detection of miRNA-21. The nanosystem showed a good selectivity towards miRNA-21 and low cytotoxicity, and its detection limit was as low as 3.8 pM. In in vivo detection, an obvious fluorescence color change from red to green can be observed in the presence of miRNA-21, as indicated by laser confocal microscope scanning. The ZnO@PDA/probe system developed in this work for miRNA-21 detection has the advantages of high sensitivity, strong specificity, low background interference, and good biocompatibility. Moreover, since the detection target of this nanosystem can be changed by simply substitute the complementary sequence of miRNA, the nanosystem can be applied in the detection of other miRNA targets. Above all, this strategy has broad application prospect in tumor diagnosis and miRNA related biology research.Author contributions
Yuanyuan Liu: investigation, methodology, validation, writing – review & editing; Ranran Wang: investigation, methodology; Fengxia Zhang: investigation, data curation; Yongshan Ma: conceptualization, resources, writing – review & editing; Tianyi Jiang: supervision, writing – original draft, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Natural Science Foundation of Shandong Provincial (ZR2021MB099), the Doctoral Research Fund of Shandong Jianzhu University (X21021Z), and State Key Laboratory of Microbial Technology Open Projects Fund (M2023-02).References
- J. V. Tricoli and J. W. Jacobson, Cancer Res., 2007, 67, 4553–4555 CrossRef CAS PubMed
.
- A. M. Mohr and J. L. Mott, Semin. Liver Dis., 2015, 35, 3–11 CrossRef CAS PubMed
.
- P. T. B. Ho, I. M. Clark and L. T. T. Le, Int. J. Mol. Sci., 2022, 23, 7167 CrossRef CAS PubMed
.
- J. J. Rossi, Cell, 2009, 137, 990–992 CrossRef CAS PubMed
.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef CAS PubMed
.
- Y. Zhang and C. Y. Zhang, Anal. Chem., 2012, 84, 224–231 CrossRef CAS PubMed
.
- H. Dong, J. Lei, H. Ju, F. Zhi, H. Wang, W. Guo, Z. Zhu and F. Yan, Angew Chem. Int. Ed. Engl., 2012, 51, 4607–4612 CrossRef CAS PubMed
.
- Y. Wang, D. Zheng, Q. Tan, M. X. Wang and L. Q. Gu, Nat. Nanotechnol., 2011, 6, 668–674 CrossRef CAS PubMed
.
- D. W. Hwang, I. C. Song, D. S. Lee and S. Kim, Small, 2010, 6, 81–88 CrossRef CAS PubMed
.
- Y. Peng, G. Yi and Z. Gao, Chem. Commun., 2010, 46, 9131–9133 RSC
.
- W. Zhou, Y. F. Tian, B. C. Yin and B. C. Ye, Anal. Chem., 2017, 89, 6120–6128 CrossRef CAS PubMed
.
- T. C. de Bang, P. Shah, S. K. Cho, S. W. Yang and S. Husted, Anal. Chem., 2014, 86, 6823–6826 CrossRef CAS PubMed
.
- W. Pan, T. Zhang, H. Yang, W. Diao, N. Li and B. Tang, Anal. Chem., 2013, 85, 10581–10588 CrossRef CAS PubMed
.
- L. Li, J. Feng, H. Liu, Q. Li, L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940–1945 RSC
.
- E. M. McConnell, I. Cozma, Q. Mou, J. D. Brennan, Y. Lu and Y. Li, Chem. Soc. Rev., 2021, 50, 8954–8994 RSC
.
- S. W. Santoro and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4262–4266 CrossRef CAS PubMed
.
- K. Kappaun, A. R. Piovesan, C. R. Carlini and R. Ligabue-Braun, J. Adv. Res., 2018, 13, 3–17 CrossRef CAS PubMed
.
- J. Liu, A. K. Brown, X. Meng, D. M. Cropek, J. D. Istok, D. B. Watson and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2056–2061 CrossRef CAS PubMed
.
- Y. L. Jing Li, J. Am. Chem. Soc., 2000, 122, 10466–10467 CrossRef
.
- S. F. Torabi, P. Wu, C. E. McGhee, L. Chen, K. Hwang, N. Zheng, J. Cheng and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 5903–5908 CrossRef CAS PubMed
.
- R. J. Lake, Z. Yang, J. Zhang and Y. Lu, Acc. Chem. Res., 2019, 52, 3275–3286 CrossRef CAS PubMed
.
- N. Carmi, S. R. Balkhi and R. R. Breaker, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 2233–2237 CrossRef CAS PubMed
.
- T. Yu, W. Zhou and J. Liu, Chembiochem, 2018, 19, 1012–1017 CrossRef CAS PubMed
.
- R. R. Breaker, Nature, 2004, 432, 838–845 CrossRef CAS PubMed
.
- Y. Huang, Y. Ma, Y. Chen, X. Wu, L. Fang, Z. Zhu and C. J. Yang, Anal. Chem., 2014, 86, 11434–11439 CrossRef CAS PubMed
.
- X. H. Zhao, L. Gong, X. B. Zhang, B. Yang, T. Fu, R. Hu, W. Tan and R. Yu, Anal. Chem., 2013, 85, 3614–3620 CrossRef CAS PubMed
.
- L. M. Lu, X. B. Zhang, R. M. Kong, B. Yang and W. Tan, J. Am. Chem. Soc., 2011, 133, 11686–11691 CrossRef CAS PubMed
.
- L. Qu, M. M. Ali, S. D. Aguirre, H. Liu, Y. Jiang and Y. Li, PLoS One, 2014, 9, e115640 CrossRef PubMed
.
- Y. Wen, Y. Xu, X. Mao, Y. Wei, H. Song, N. Chen, Q. Huang, C. Fan and D. Li, Anal. Chem., 2012, 84, 7664–7669 CrossRef CAS PubMed
.
- B. Li, J. Liu and H. Zhou, Anal. Chim. Acta, 2018, 1008, 96–102 CrossRef CAS PubMed
.
- F. Wang, L. Freage, R. Orbach and I. Willner, Anal. Chem., 2013, 85, 8196–8203 CrossRef CAS PubMed
.
- F. Wang, J. Elbaz, C. Teller and I. Willner, Angew Chem. Int. Ed. Engl., 2011, 50, 295–299 CrossRef CAS PubMed
.
- J. Elbaz, M. Moshe, B. Shlyahovsky and I. Willner, Chemistry, 2009, 15, 3411–3418 CrossRef CAS PubMed
.
- X. Liu, Y. Wang, C. Y. Effah, L. Wu, F. Yu, J. Wei, G. Mao, Y. Xiong and L. He, Talanta, 2022, 243, 123377 CrossRef CAS PubMed
.
- W. Ma, Y. Zhan, Y. Zhang, C. Mao, X. Xie and Y. Lin, Signal Transduction Targeted Ther., 2021, 6, 351 CrossRef CAS PubMed
.
- R. Kumarswamy, I. Volkmann and T. Thum, RNA Biol., 2011, 8, 706–713 CrossRef CAS PubMed
.
- J. T. Yi, T. T. Chen, J. Huo and X. Chu, Anal. Chem., 2017, 89, 12351–12359 CrossRef CAS PubMed
.
- J. Li, W. Zheng, A. H. Kwon and Y. Lu, Nucleic Acids Res., 2000, 28, 481–488 CrossRef CAS PubMed
.
- S. Javanmardi, M. R. Aghamaali, S. S. Abolmaali, S. Mohammadi and A. M. Tamaddon, Curr. Gene Ther., 2017, 16, 375–389 CrossRef PubMed
.
- Z. Y. Zhang, Y. D. Xu, Y. Y. Ma, L. L. Qiu, Y. Wang, J. L. Kong and H. M. Xiong, Angew Chem. Int. Ed. Engl., 2013, 52, 4127–4131 CrossRef CAS PubMed
.
- K. Zhou, H. Liu, S. Zhang, X. Huang, Y. Wang, G. Huang, B. D. Sumer and J. Gao, J. Am. Chem. Soc., 2012, 134, 7803–7811 CrossRef CAS PubMed
.
- Y. Liu, K. Ai and L. Lu, Chem. Rev., 2014, 114, 5057–5115 CrossRef CAS PubMed
.
- D. He, X. He, X. Yang and H. W. Li, Chem. Sci., 2017, 8, 2832–2840 RSC
.
- D. T. Humphreys, C. J. Hynes, H. R. Patel, G. H. Wei, L. Cannon, D. Fatkin, C. M. Suter, J. L. Clancy and T. Preiss, PLoS One, 2012, 7, e30933 CrossRef CAS PubMed
.
- L. Lu, P. Schwartz, L. Scarampi, T. Rutherford, E. M. Canuto, H. Yu and D. Katsaros, Gynecol. Oncol., 2011, 122, 366–371 CrossRef CAS PubMed
.
- Y. Gao, B. Feng, S. Han, K. Zhang, J. Chen, C. Li, R. Wang and L. Chen, Cell. Physiol. Biochem., 2016, 38, 427–448 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra00636d |
| This journal is © The Royal Society of Chemistry 2024 |