Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Turmeric extract-mediated biogenic synthesis of Ag@SeO2 magnetic nanoparticles: characterization, optimization, antibacterial and antioxidant activities†
Abeer A. Ghoniema,
Khaled M. Elattar *b,
Fatimah O. Al-Otibic,
Ashraf Elsayedd,
Mohammed S. El-Hersha,
Ayman Y. El-Khateebe,
Yosra A. Helmyf and
WesamEldin I. A. Saber
*b,
Fatimah O. Al-Otibic,
Ashraf Elsayedd,
Mohammed S. El-Hersha,
Ayman Y. El-Khateebe,
Yosra A. Helmyf and
WesamEldin I. A. Saber *a
*a
aMicrobial Activity Unit, Department of Microbiology, Soils, Water and Environment Research Institute, Agricultural Research Center, Giza 12619, Egypt. E-mail: abeer.abdelkhalik@yahoo.com; m.elhersh@yahoo.com; wesameldin.saber@arc.sci.eg
bUnit of Genetic Engineering and Biotechnology, Faculty of Science, Mansoura University, El-Gomhoria St., Mansoura, 35516, Egypt. E-mail: khaledelattar2@yahoo.com
cBotany and Microbiology Department, Faculty of Science, King Saud University, Riyadh 11451, Saudi Arabia. E-mail: falotibi@ksu.edu.sa
dBotany Department, Faculty of Science, Mansoura University, El-Gomhoria St., Mansoura 35516, Egypt. E-mail: ashraf-badawy@mans.edu.eg
eAgricultural Chemistry Department, Faculty of Agriculture, Mansoura University, El-Gomhoria St., Mansoura 35516, Egypt. E-mail: aymanco@mans.edu.eg
fDepartment of Veterinary Science, Martin-Gatton College of Agriculture, Food, and Environment, University of Kentucky, Lexington, KY 40546, USA. E-mail: yosra.helmy@uky.edu
First published on 27th February 2024
Abstract
This study bio-synthesized Ag@SeO2 bmNPs successfully, using turmeric ethanol extract, and characterized them using various techniques. The FT-IR analysis reveals the involvement of these plant-derived compounds, especially phenolics, in the reduction process by acting as electron donors and stabilizing/capping agents. Zeta potential analysis showed a slight negative surface charge for the stability of Ag@SeO2 NPs, where TEM revealed spherical nanoparticles with an average size of 20 nm. The XRD confirmed crystallinity and a core–shell structure, and EDX identified elements consistent with Ag@SeO2 and a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Ag/Se atomic ratio. Further, SEM supported the spherical shape and uniform size. These findings highlight the successful biosynthesis of Ag@SeO2 bmNPs with promising properties for diverse applications. Moreover, the Box–Behnken design (BBD) and artificial neural network (ANN) model were engaged to optimize Ag@SeO2 bmNP biosynthesis. BBD identified significant influences of pH, bioconversion temperature, time, and turmeric concentration on bmNP yield, with adjusted R2 and predictive R2 being 0.9075 and 0.8829, respectively. However, its limitations were revealed by a significant lack of fit. ANN modeling with a 3–5–7–1 topology showed superior predictive accuracy and identified optimal conditions for maximizing yield (pH 9.83, 51.7 °C, 1.0 h, 3.71 mg mL−1 turmeric). Validation experiments confirmed the model's reliability. Turmeric extract exhibited significantly higher amounts of phenolics, and flavonoids compared to the bmNPs, suggesting its potential for strong antioxidant activity. Both turmeric extract and bmNPs displayed antioxidant activity in ABTS and DPPH assays, with turmeric extract being the most potent due to its curcuminoid content. The potential activity of Ag@SeO2 bmNPs against S. aureus, K. pneumonia, E. coli, and B. cereus was investigated, with inhibition zones ranging from 22 to 32 mm. The MIC values of tested NPs towards pathogenic bacteria ranged from 165.625 and 331.25 μg mL−1.
1 Ag/Se atomic ratio. Further, SEM supported the spherical shape and uniform size. These findings highlight the successful biosynthesis of Ag@SeO2 bmNPs with promising properties for diverse applications. Moreover, the Box–Behnken design (BBD) and artificial neural network (ANN) model were engaged to optimize Ag@SeO2 bmNP biosynthesis. BBD identified significant influences of pH, bioconversion temperature, time, and turmeric concentration on bmNP yield, with adjusted R2 and predictive R2 being 0.9075 and 0.8829, respectively. However, its limitations were revealed by a significant lack of fit. ANN modeling with a 3–5–7–1 topology showed superior predictive accuracy and identified optimal conditions for maximizing yield (pH 9.83, 51.7 °C, 1.0 h, 3.71 mg mL−1 turmeric). Validation experiments confirmed the model's reliability. Turmeric extract exhibited significantly higher amounts of phenolics, and flavonoids compared to the bmNPs, suggesting its potential for strong antioxidant activity. Both turmeric extract and bmNPs displayed antioxidant activity in ABTS and DPPH assays, with turmeric extract being the most potent due to its curcuminoid content. The potential activity of Ag@SeO2 bmNPs against S. aureus, K. pneumonia, E. coli, and B. cereus was investigated, with inhibition zones ranging from 22 to 32 mm. The MIC values of tested NPs towards pathogenic bacteria ranged from 165.625 and 331.25 μg mL−1.
1. Introduction
Biogenic nanoparticles (NPs) are nanoparticles that are synthesized using biological resources, such as plants, bacteria, and fungi. Biogenic NPs have several advantages over chemically synthesized NPs, including their low cost, ease of synthesis, and biocompatibility.1,2 Turmeric extract is a rich source of curcumin, a bioactive compound with a wide range of biological activities, including antioxidant, anti-inflammatory, and anticancer activities.3–6 Curcumin has also been shown to reduce the toxicity of silver nanoparticles (Ag-NPs) as well as the expected improved antibacterial activity owing to the synergistic effects of Ag, Se, and curcumin.7Selenium dioxide (SeO2) is a semiconductor material with a wide range of applications in catalysis, electronics, and biomedicine.8 SeO2 NPs have been shown to exhibit excellent antifungal and antioxidant activities.9,10 The bmNPs have the potential to be used in a variety of applications, including biomedical applications for developing new drugs and diagnostic tools for the treatment and detection of fungal infections and other diseases.11 The bmNPs could be used in the development of new water purification and wastewater treatment technologies.12 The bmNPs could be used in the development of new food packaging materials and antimicrobial coatings to prevent food spoilage.13
Plant-mediated synthesis of metal nanoparticles (PMSN) is a green and sustainable approach to producing metal nanoparticles using plant extracts. It has emerged as a promising alternative to traditional physical and chemical methods, which are often energy-intensive, expensive, and environmentally unfavorable.14 PMSN harnesses the unique properties of plant extracts to reduce and stabilize metal ions into nanoparticles. Plant extracts contain a variety of phytochemicals, such as alkaloids, flavonoids, terpenoids, and polysaccharides, which can act as reducing agents, capping agents, and stabilizers.15,16 These phytochemicals can also control the size, shape, and properties of the synthesized nanoparticles.
The optimization of metallic nanoparticle synthesis is a critical facet involving the meticulous identification and adjustment of reaction parameters to achieve nanoparticles possessing the desired size, shape, and properties.17 Diverse factors exert influence throughout this process; notably, precursor salt concentration plays a pivotal role, impacting both nanoparticle size and nucleation rate.18 The nature and concentration of the reducing agent are significant determinants affecting the resulting size, shape, and properties of the nanoparticles, whereas capping agents serve the crucial role of stabilizing nanoparticles and preventing aggregation.19,20 Reaction temperature modulates nucleation and growth rates, while pH exerts influence over precursor salt solubility and nanoparticle stability.21 Although one-at-a-time optimization, albeit straightforward, proves time-intensive and may not ascertain the true optimum,22 the experimental design approach of response surface methodology permits the simultaneous statistical analysis of multiple parameters, revealing intricate interactions.23 Furthermore, the integration of machine learning facilitates the prediction of nanoparticle properties based on synthesis parameters, thereby guiding the optimization process.24 The refinement of synthesis is exemplified by the controlled modulation of the size and nucleation rate of silver nanoparticles through the adjustment of precursor salt concentration.25 Recognizing the pivotal role of optimization in tailoring metallic nanoparticles with specific characteristics for diverse applications, scientists rigorously control synthesis conditions to ensure precise outcomes.26,27
In the burgeoning field of nanotechnology, biological researchers are trying to unveil a pioneering application of artificial intelligence in the modeling of biosynthesis for nanometals, and leveraging the unique capacity of artificial neural networks (ANNs) to capture temporal dependencies and intricate sequential patterns. Our study presents a methodological framework that brings unprecedented insight into the dynamic molecular processes involved.
The advantages of employing ANNs become evident as they outperform traditional modeling approaches. The benefits of employing ANNs range from their ability to discern complex biosynthetic pathways to predicting optimal conditions for enhanced efficiency. The iterative nature of ANNs mirrors the iterative intricacies of biosynthesis, enabling a more nuanced and precise representation of the intricate interplay of biological components.28,29 This research sheds light on the seamless integration of ANNs as a transformative tool, offering a data-driven lens that not only elucidates the biosynthesis of nonmetals but also opens new avenues for optimizing and advancing nanometal production.
Turmeric ethanol extract is a popular herbal supplement that is made by soaking turmeric rhizomes in ethanol. Ethanol is a good solvent for extracting active phytochemicals from turmeric, including curcuminoids, volatile oils, and other phenolic compounds.30 Curcuminoids are the most active phytochemical components of turmeric ethanol extract.31 Curcuminoids are polyphenolic compounds that have been shown to have a widespread variety of biological activities, comprising antioxidant, anti-inflammatory, and anticancer features.32 The three main curcuminoids are curcumin, demethoxy-curcumin, and bis-demethoxy-curcumin.33 Curcumin is the most abundant and well-studied curcuminoid.34 Volatile oils are another important class of phytochemicals in turmeric ethanol extract. Volatile oils are liable for the pungent aroma of turmeric.35
The main volatile oils in turmeric are turmerone, ar-turmerone, and zingiberene. Volatile oils have been shown to have antibacterial, antifungal, and insecticidal properties.36 Other phenolic compounds in turmeric ethanol extract include ar-curcumin, ferulic acid, and caffeic acid. These compounds also displayed a variety of biological features, including antioxidant, anti-inflammatory, and anticancer features.37 Turmeric ethanol extract presents a wide assortment of potential health benefits as it is considered to reduce inflammation, boost the immune system, protect against heart disease, prevent cancer, improve brain function, relieve pain and arthritis symptoms, protect the liver, and improve digestion.38,39 Turmeric ethanol extract is generally safe for most people to take. Turmeric extracts protect the brain from damage caused by neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease.40 Turmeric extracts have also been shown to protect the heart from damage caused by heart disease. This is due to their ability to improve cholesterol levels, reduce inflammation, and prevent blood clots.41 In addition to these biological activities, turmeric extracts have also been shown to have beneficial effects on digestion, liver health, and mood.42
In this study, we report the green synthesis and characterization of Ag–SeO2 core/shell magnetic nanoparticles using turmeric extract as a reducing and stabilizing agent. We also investigate the optimization of the synthesis process, phytochemical analyses, and the antibacterial and antioxidant activities of the turmeric extract and Ag–SeO2 bmNPs.
2. Materials and methods
2.1. Instruments
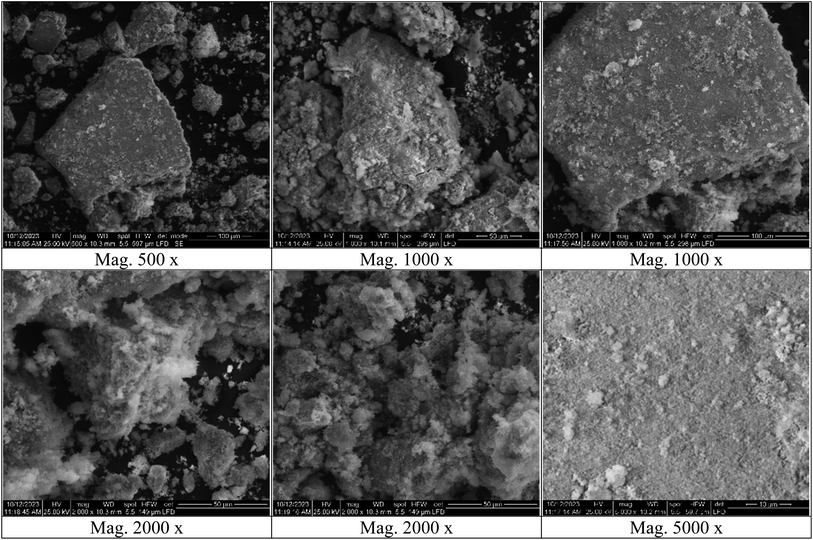
Fourier-transform infrared spectroscopy (FT-IR) analysis was performed on a Thermo-Fisher Nicolet IS10 Spectrophotometer in the frequency range of 500–4000 cm−1. Zetasizer and zeta potential analyses were performed on HORIBA Scientific SZ-100 “Ver 2.40”. The surface features, shape, and elemental composition of the silver-selenium dioxide nanocomposite were analyzed using scanning electron microscopy (SEM) and energy dispersive X-ray spectroscopy (EDX) on an FEI Czech SEM-type instrument at an accelerating voltage of 25 kV. Transmission electron microscopy (TEM) analyses were performed on a ThermoScientific Talos F200i instrument using a carbon-coated grid. X-ray diffraction (XRD) analysis was performed on a Pan Analytical Philips instrument to determine the material type, phase, crystallographic structure, and physical properties of the nanocomposite.2.2. Preparation of turmeric extract
Turmeric (Curcuma longa) powder was purchased from the local market in Mansoura city, Egypt. A weighed 10 grams of turmeric powder was placed in a clean conical flask (250 mL). 100 mL of 80% ethanol was added to the plant powder and the mixture was stirred to ensure that all of the turmeric powder was wetted by the ethanol. The flask was covered with a lid or parafilm and placed in a dark at 25 °C. The mixture was soaked for 24 hours and stirred occasionally.43 The mixture was then filtered with a filter paper and the filtrate was used immediately. The turmeric extract was stored in a cool, and dark place to preserve its quality.2.3. Biosynthesis and storage of Ag@SeO2 nanocomposite
2.4. Optimization Ag@SeO2 biosynthesis conditions
According to BBD, each of the four factors was examined at low, middle, and high levels. Thus, a four-factor matrix of various combinations of experimental runs was generated, composed of 27 runs as shown in Table 1. The design was experimentally repeated to ensure accuracy, and each run was carried out in triplicates. Once performing the laboratory experiments, the biosynthesized Ag@SeO2 bmNPs were determined in response to each run condition. The laboratory-recorded data were statistically analyzed to define the significant factors and the goodness of the BBD model, in this connection, the analysis of variance (ANOVA) and determination coefficient (R2) were calculated. Furthermore, the association between the four tested variables and Ag@SeO2 bmNPs biosynthesis, as well as the prediction of the optimum level of each variable were elucidated. The function of the second-order polynomial quadratic model (eqn (1)) was applied:
| Y = β0 + ∑βiXi + ∑βijXiXj + βiiXi2 | (1) |
| Run | Investigated variablea | Ag@SeO2 bmNP (μg mL−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Actual | Box–Behnken | ANNb | ||||||||
| X1 | X2 | X3 | X4 | Predicted | Error | Predicted | Error | Validation | ||
| a X1; pH, X2; bioconversion temperature (°C), X3; bioconversion time (h), X4; concentration of turmeric (mg mL−1).b For ANN, the training group was composed of 54, and the validation dataset was composed of 27 runs. | ||||||||||
| 1 | 4 | 40 | 3 | 8.047 | 456.86 | 417.05 | 39.81 | 452.32 | 4.54 | Validation |
| 2 | 10 | 40 | 3 | 8.047 | 413.65 | 402.16 | 11.49 | 412.78 | 0.87 | Validation |
| 3 | 4 | 80 | 3 | 8.047 | 418.30 | 400.82 | 17.48 | 417.23 | 1.07 | Training |
| 4 | 10 | 80 | 3 | 8.047 | 403.44 | 414.18 | −10.74 | 397.29 | 6.15 | Validation |
| 5 | 7 | 60 | 1 | 3.714 | 932.40 | 866.21 | 66.19 | 926.87 | 5.53 | Validation |
| 6 | 7 | 60 | 5 | 3.714 | 753.55 | 663.56 | 89.99 | 748.66 | 4.89 | Validation |
| 7 | 7 | 60 | 1 | 12.380 | 206.80 | 267.35 | −60.55 | 206.39 | 0.41 | Validation |
| 8 | 7 | 60 | 5 | 12.380 | 173.87 | 210.97 | −37.10 | 171.23 | 2.64 | Training |
| 9 | 4 | 60 | 3 | 3.714 | 872.98 | 859.65 | 13.33 | 873.03 | −0.05 | Validation |
| 10 | 10 | 60 | 3 | 3.714 | 843.30 | 843.08 | 0.22 | 841.98 | 1.32 | Validation |
| 11 | 4 | 60 | 3 | 12.380 | 280.59 | 318.11 | −37.52 | 274.32 | 6.27 | Training |
| 12 | 10 | 60 | 3 | 12.380 | 282.50 | 333.16 | −50.66 | 283.80 | −1.30 | Training |
| 13 | 7 | 40 | 1 | 8.047 | 441.89 | 389.16 | 52.73 | 438.73 | 3.16 | Validation |
| 14 | 7 | 80 | 1 | 8.047 | 443.58 | 384.51 | 59.07 | 446.96 | −3.38 | Validation |
| 15 | 7 | 40 | 5 | 8.047 | 160.40 | 257.09 | −96.69 | 168.75 | −8.35 | Validation |
| 16 | 7 | 80 | 5 | 8.047 | 167.25 | 257.54 | −90.29 | 172.06 | −4.81 | Training |
| 17 | 4 | 60 | 1 | 8.047 | 573.93 | 657.41 | −83.48 | 576.78 | −2.85 | Validation |
| 18 | 10 | 60 | 1 | 8.047 | 442.37 | 480.20 | −37.83 | 444.61 | −2.24 | Training |
| 19 | 4 | 60 | 5 | 8.047 | 397.59 | 351.45 | 46.14 | 397.97 | −0.38 | Training |
| 20 | 10 | 60 | 5 | 8.047 | 601.00 | 527.13 | 73.87 | 612.51 | −11.51 | Validation |
| 21 | 7 | 40 | 3 | 3.714 | 756.90 | 849.42 | −92.52 | 752.61 | 4.29 | Training |
| 22 | 7 | 80 | 3 | 3.714 | 421.35 | 489.37 | −68.02 | 419.29 | 2.06 | Validation |
| 23 | 7 | 40 | 3 | 12.380 | 57.00 | −34.26 | 91.26 | 55.61 | 1.39 | Training |
| 24 | 7 | 80 | 3 | 12.380 | 422.59 | 321.59 | 101.00 | 425.11 | −2.52 | Training |
| 25 | 7 | 60 | 3 | 8.047 | 395.07 | 414.82 | −19.75 | 415.49 | −20.42 | Training |
| 26 | 7 | 60 | 3 | 8.047 | 388.56 | 414.82 | −26.26 | 415.49 | −26.93 | Validation |
| 27 | 7 | 60 | 3 | 8.047 | 443.09 | 414.82 | 28.27 | 415.49 | 27.60 | Training |
| 28 | 4 | 40 | 3 | 8.047 | 445.85 | 417.05 | 28.80 | 452.32 | −6.47 | Training |
| 29 | 10 | 40 | 3 | 8.047 | 401.62 | 402.16 | −0.54 | 412.78 | −11.16 | Validation |
| 30 | 4 | 80 | 3 | 8.047 | 406.50 | 400.82 | 5.68 | 417.23 | −10.73 | Training |
| 31 | 10 | 80 | 3 | 8.047 | 397.41 | 414.18 | −16.77 | 397.29 | 0.12 | Training |
| 32 | 7 | 60 | 1 | 3.714 | 915.20 | 866.21 | 48.99 | 926.87 | −11.67 | Training |
| 33 | 7 | 60 | 5 | 3.714 | 747.52 | 663.56 | 83.96 | 748.66 | −1.14 | Training |
| 34 | 7 | 60 | 1 | 12.380 | 198.40 | 267.35 | −68.95 | 206.39 | −7.99 | Training |
| 35 | 7 | 60 | 5 | 12.380 | 162.81 | 210.97 | −48.16 | 171.23 | −8.42 | Training |
| 36 | 4 | 60 | 3 | 3.714 | 866.94 | 859.65 | 7.29 | 873.03 | −6.09 | Validation |
| 37 | 10 | 60 | 3 | 3.714 | 835.10 | 843.08 | −7.98 | 841.98 | −6.88 | Validation |
| 38 | 4 | 60 | 3 | 12.380 | 269.54 | 318.11 | −48.57 | 274.32 | −4.78 | Training |
| 39 | 10 | 60 | 3 | 12.380 | 275.21 | 333.16 | −57.95 | 283.80 | −8.59 | Validation |
| 40 | 7 | 40 | 1 | 8.047 | 435.85 | 389.16 | 46.69 | 438.73 | −2.88 | Validation |
| 41 | 7 | 80 | 1 | 8.047 | 433.45 | 384.51 | 48.94 | 446.96 | −13.51 | Training |
| 42 | 7 | 40 | 5 | 8.047 | 145.70 | 257.09 | −111.39 | 168.75 | −23.05 | Training |
| 43 | 7 | 80 | 5 | 8.047 | 161.54 | 257.54 | −96.00 | 172.06 | −10.52 | Training |
| 44 | 4 | 60 | 1 | 8.047 | 573.93 | 657.41 | −83.48 | 576.78 | −2.85 | Training |
| 45 | 10 | 60 | 1 | 8.047 | 435.35 | 480.20 | −44.85 | 444.61 | −9.26 | Training |
| 46 | 4 | 60 | 5 | 8.047 | 389.45 | 351.45 | 38.00 | 397.97 | −8.52 | Training |
| 47 | 10 | 60 | 5 | 8.047 | 601.00 | 527.13 | 73.87 | 612.51 | −11.51 | Training |
| 48 | 7 | 40 | 3 | 3.714 | 748.60 | 849.42 | −100.82 | 752.61 | −4.01 | Validation |
| 49 | 7 | 80 | 3 | 3.714 | 421.35 | 489.37 | −68.02 | 419.29 | 2.06 | Training |
| 50 | 7 | 40 | 3 | 12.380 | 57.00 | −34.26 | 91.26 | 55.61 | 1.39 | Training |
| 51 | 7 | 80 | 3 | 12.380 | 414.63 | 321.59 | 93.04 | 425.11 | −10.48 | Validation |
| 52 | 7 | 60 | 3 | 8.047 | 388.32 | 414.82 | −26.50 | 415.49 | −27.17 | Training |
| 53 | 7 | 60 | 3 | 8.047 | 367.63 | 414.82 | −47.19 | 415.49 | −47.86 | Training |
| 54 | 7 | 60 | 3 | 8.047 | 438.67 | 414.82 | 23.85 | 415.49 | 23.18 | Training |
| 55 | 4 | 40 | 3 | 8.047 | 467.87 | 417.05 | 50.82 | 452.32 | 15.55 | Training |
| 56 | 10 | 40 | 3 | 8.047 | 425.68 | 402.16 | 23.52 | 412.78 | 12.90 | Training |
| 57 | 4 | 80 | 3 | 8.047 | 430.40 | 400.82 | 29.58 | 417.23 | 13.17 | Training |
| 58 | 10 | 80 | 3 | 8.047 | 409.46 | 414.18 | −4.72 | 397.29 | 12.17 | Validation |
| 59 | 7 | 60 | 1 | 3.714 | 949.60 | 866.21 | 83.39 | 926.87 | 22.73 | Training |
| 60 | 7 | 60 | 5 | 3.714 | 759.57 | 663.56 | 96.01 | 748.66 | 10.91 | Training |
| 61 | 7 | 60 | 1 | 12.380 | 214.11 | 267.35 | −53.24 | 206.39 | 7.72 | Training |
| 62 | 7 | 60 | 5 | 12.380 | 184.89 | 210.97 | −26.08 | 171.23 | 13.66 | Training |
| 63 | 4 | 60 | 3 | 3.714 | 878.96 | 859.65 | 19.31 | 873.03 | 5.93 | Training |
| 64 | 10 | 60 | 3 | 3.714 | 851.20 | 843.08 | 8.12 | 841.98 | 9.22 | Training |
| 65 | 4 | 60 | 3 | 12.380 | 291.63 | 318.11 | −26.48 | 274.32 | 17.31 | Validation |
| 66 | 10 | 60 | 3 | 12.380 | 289.63 | 333.16 | −43.53 | 283.80 | 5.83 | Training |
| 67 | 7 | 40 | 1 | 8.047 | 447.94 | 389.16 | 58.78 | 438.73 | 9.21 | Validation |
| 68 | 7 | 80 | 1 | 8.047 | 453.67 | 384.51 | 69.16 | 446.96 | 6.71 | Training |
| 69 | 7 | 40 | 5 | 8.047 | 175.80 | 257.09 | −81.29 | 168.75 | 7.05 | Training |
| 70 | 7 | 80 | 5 | 8.047 | 173.42 | 257.54 | −84.12 | 172.06 | 1.36 | Training |
| 71 | 4 | 60 | 1 | 8.047 | 586.51 | 657.41 | −70.90 | 576.78 | 9.73 | Training |
| 72 | 10 | 60 | 1 | 8.047 | 449.52 | 480.20 | −30.68 | 444.61 | 4.91 | Training |
| 73 | 4 | 60 | 5 | 8.047 | 405.64 | 351.45 | 54.19 | 397.97 | 7.67 | Training |
| 74 | 10 | 60 | 5 | 8.047 | 642.23 | 527.13 | 115.10 | 612.51 | 29.72 | Training |
| 75 | 7 | 40 | 3 | 3.714 | 764.50 | 849.42 | −84.92 | 752.61 | 11.89 | Validation |
| 76 | 7 | 80 | 3 | 3.714 | 394.87 | 489.37 | −94.50 | 419.29 | −24.42 | Training |
| 77 | 7 | 40 | 3 | 12.380 | 38.75 | −34.26 | 73.01 | 55.61 | −16.86 | Training |
| 78 | 7 | 80 | 3 | 12.380 | 430.82 | 321.59 | 109.23 | 425.11 | 5.71 | Training |
| 79 | 7 | 60 | 3 | 8.047 | 453.06 | 414.82 | 38.24 | 415.49 | 37.57 | Training |
| 80 | 7 | 60 | 3 | 8.047 | 432.64 | 414.82 | 17.82 | 415.49 | 17.15 | Validation |
| 81 | 7 | 60 | 3 | 8.047 | 426.33 | 414.82 | 11.51 | 415.49 | 10.84 | Training |
Data handling employed a three-pronged approach. The first dataset served as the training ground, where backpropagation optimized network parameters by minimizing error at each neuron. This facilitated robust learning and initial weight establishment. The second dataset fulfilled a validation role, allowing for training interruptions and optimal model selection to prevent overfitting. Finally, a dedicated “outer” dataset held back from training assessed the model's true predictive strength in a final evaluation step, ensuring independent and rigorous performance benchmarks.
The model architecture revolved around a three-layered setup; an input layer, a hidden layer (or multiple layers as determined), and an output layer. Four input neurons mapped directly to the experimental parameters – pH, bioconversion temperature, time, and turmeric concentration. The single output neuron represented the predicted Ag@SeO2 bio-nanoparticle biosynthesis. Determining the optimal hidden layer configuration involved an iterative process.
For effective Ag@SeO2 bio-nanoparticle biosynthesis modeling, the data were strategically and randomly partitioned into training and validation datasets employing a holdback propagation ratio of 0.3333, indicating that approximately one-third of the data was reserved for validation purposes for rigorous validation. The training dataset comprised 54 runs, which were utilized to minimize prediction error and establish neural weights. Subsequently, a separate validation dataset consisting of 27 runs was employed to monitor the performance of the ANN during training and to determine the optimal stopping point, thereby selecting the finest model. This approach ensured that the ANN did not over fit the training data and generalized well to unseen conditions. Additionally, a third external dataset was reserved for testing the robustness of the ANN model and evaluating its predictive competencies on independent data. To prevent overfitting and improve the generalization of the model, the regularization technique was employed, including the squared penalty method. This method penalizes overly complex models by adding a regularization term to the loss function, encouraging the ANN to prioritize simpler solutions that generalize better to new data.
During training, a learning rate of 0.1 was utilized to facilitate efficient convergence and optimal performance of the ANN model. This learning rate was determined through trial-and-error testing across 5000 training iterations per phase, where various ANN parameters such as the learning rate and hidden layer neuron count were optimized. This meticulous approach yielded the ideal network structure for the best model, ensuring robust performance in predicting Ag@SeO2 bio-nanoparticle biosynthesis.
Training continued until a combination of minimal error, including root average squared error and mean absolute deviation, accompanied by maximized R-squared values, was achieved. These performance metrics signified the peak performance of Ag@SeO2 bio-nanoparticle biosynthesis prediction, with model outputs closely resembling actual experimental values. The network's predictive accuracy was rigorously assessed by comparing its predictions with experimental data and evaluating its ability to generalize to unseen samples, thereby ensuring the reliability and validity of the ANN model.
2.5. Phytochemical analyses
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) according to the manufacturer's instructions. The FC reagent (5 mL) was added to each tube containing the sample (100 μL) and mixed gently. After 3 minutes, sodium carbonate solution (4 mL, 75 g L−1) was added and the mixture was well-mixed. The final volume was adjusted by distilled water to 10 mL. The tubes were incubated in the dark at 40 °C for 30 minutes. This allows the phenolics in the samples to react with the FC reagent and develop a blue color. A blank was prepared using the same previous steps but without any sample material. A standard curve was prepared from a series of standard (reference) solutions using gallic acid with concentrations ranging from 0 to 200 μg mL−1. After incubation, the absorbance of each tube was measured at a wavelength of 765 nm.45 The gallic acid standard curve (y = 0.0062x, r2 = 0.987) was applied to calculate the phenolic contents as milligrams of gallic acid equivalents per gram of sample (mg GAE per g).
9) according to the manufacturer's instructions. The FC reagent (5 mL) was added to each tube containing the sample (100 μL) and mixed gently. After 3 minutes, sodium carbonate solution (4 mL, 75 g L−1) was added and the mixture was well-mixed. The final volume was adjusted by distilled water to 10 mL. The tubes were incubated in the dark at 40 °C for 30 minutes. This allows the phenolics in the samples to react with the FC reagent and develop a blue color. A blank was prepared using the same previous steps but without any sample material. A standard curve was prepared from a series of standard (reference) solutions using gallic acid with concentrations ranging from 0 to 200 μg mL−1. After incubation, the absorbance of each tube was measured at a wavelength of 765 nm.45 The gallic acid standard curve (y = 0.0062x, r2 = 0.987) was applied to calculate the phenolic contents as milligrams of gallic acid equivalents per gram of sample (mg GAE per g).2.6. Potential antioxidant activity
| Inhibition (%) = [(Abs.(control) − Abs.(sample))/Abs.(control)] × 100 | (2) |
The antioxidant activity can also be expressed as an IC50 value, which is the concentration of the antioxidant sample required to inhibit the ABTS radical by 50%.
| DPPH scavenging activity (%) = [(Abs.(control) − Abs.(sample))/Abs.(control)] × 100 | (3) |
2.7. Antibacterial activity
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 to 82.8125 μg mL−1. The control consisted of solely inoculated broth and was incubated alongside the dilutions for 24 hours at 37 °C. The absence of visible growth in a tube defined the MIC endpoint. This finding was validated by visually comparing turbidity before and after incubation and further supported by measuring the OD at λ = 600 nm.50
200 to 82.8125 μg mL−1. The control consisted of solely inoculated broth and was incubated alongside the dilutions for 24 hours at 37 °C. The absence of visible growth in a tube defined the MIC endpoint. This finding was validated by visually comparing turbidity before and after incubation and further supported by measuring the OD at λ = 600 nm.503. Results and discussion
3.1. Characterization of the nanocomposite
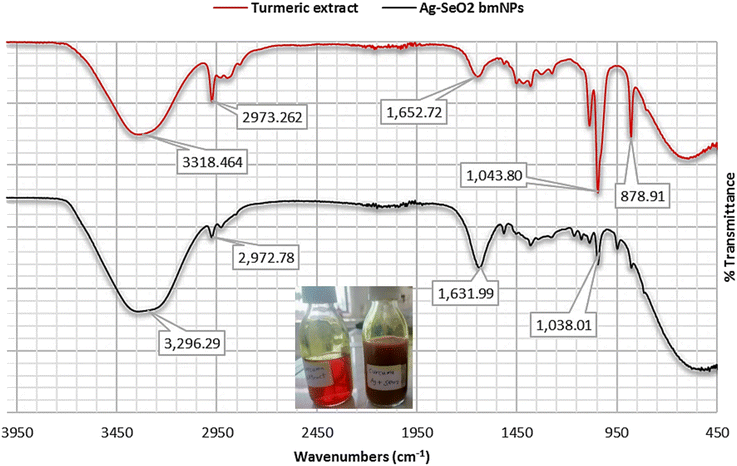
On the other hand, the FT-IR spectrum of Ag@SeO2 bmNPs revealed that broader O–H stretching vibrations at 3338 and 3296 cm−1, compared to the extract, suggest interactions between the hydroxyl groups and the surface functional groups of the nanoparticles. The presence of aliphatic groups in the nanoparticles is confirmed by the similar absorption band at 2972 cm−1, indicating they remain after bio-synthesis. A slight shift in the 2148 cm−1 absorption band further supports the notion of interactions between the biomolecules and the nanoparticles' surface. The shifted C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching absorption band at 1631 cm−1 hints at potential coordination between these groups and metal ions on the nanoparticles' surface. A new absorption band at 1515 cm−1 suggests adsorption on the nanoparticles, indicating they interact with various biomolecules. Just like in the extract, the 1381 cm−1 absorption band confirms the presence of aromatic CH groups in the nanoparticles. Similar to the extract, the absorption bands at 1038 and 1045 cm−1 suggest that carbohydrates/polysaccharides are still present in the nanoparticles. The exciting new absorption band at 947 cm−1 potentially arises from Se–O stretching vibrations, confirming the presence of SeO2 in the bio-synthesized nanoparticles. Stretching vibrations between metal (Se) and oxygen atoms might be responsible for the absorption band at 486 cm−1, further supporting the presence of the desired nanoparticles.
O stretching absorption band at 1631 cm−1 hints at potential coordination between these groups and metal ions on the nanoparticles' surface. A new absorption band at 1515 cm−1 suggests adsorption on the nanoparticles, indicating they interact with various biomolecules. Just like in the extract, the 1381 cm−1 absorption band confirms the presence of aromatic CH groups in the nanoparticles. Similar to the extract, the absorption bands at 1038 and 1045 cm−1 suggest that carbohydrates/polysaccharides are still present in the nanoparticles. The exciting new absorption band at 947 cm−1 potentially arises from Se–O stretching vibrations, confirming the presence of SeO2 in the bio-synthesized nanoparticles. Stretching vibrations between metal (Se) and oxygen atoms might be responsible for the absorption band at 486 cm−1, further supporting the presence of the desired nanoparticles.
By analyzing these absorption bands, we gain valuable insights into the molecular structure of both the turmeric extract and the bio-synthesized Ag@SeO2 nanoparticles. The FT-IR spectra confirm the presence of functional groups associated with biomolecules like phenolics, carbohydrates, and proteins in both the turmeric extract and the Ag@SeO2 bmNPs. The broader and shifted absorption bands in the bmNPs spectrum compared to the extract suggest interactions between the biomolecules and the nanoparticles' surface.51 These interactions could involve hydrogen bonding, electrostatic interactions, or coordination with metal ions, potentially contributing to the bio-reduction and stabilization of the nanoparticles.52 The presence of new absorption bands in the bmNPs spectrum, like the amide band and the Se–O stretching vibration, further supports the hypothesis of biomolecule adsorption and SeO2 formation on the nanoparticle surface.
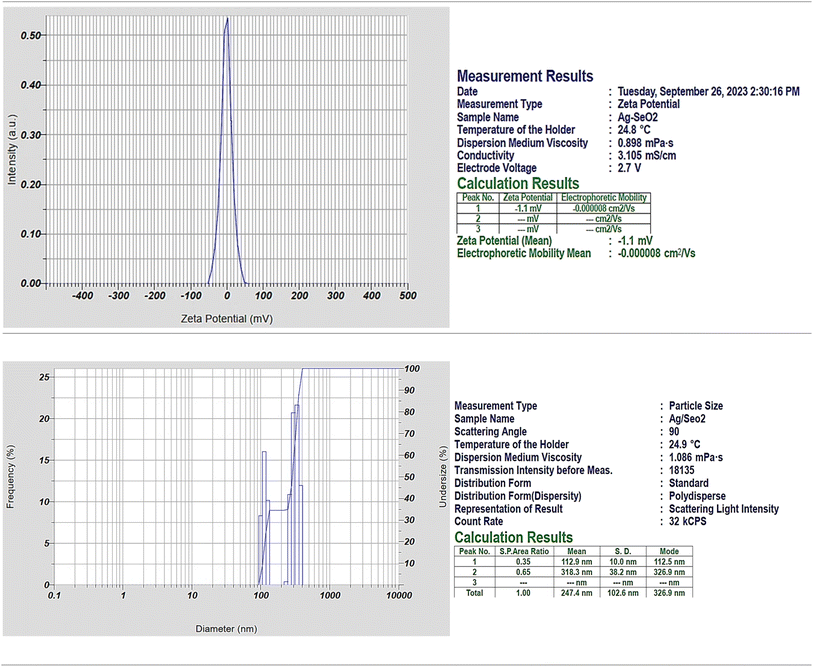
The negative electrophoretic mobility indicates that the nanoparticles are moving in the opposite direction of the electric field, which is consistent with the fact that they have a negative surface charge (Fig. 2). Particularly, the zeta potential results suggest that the Ag@SeO2 bmNPs have a slightly negative surface charge and are stable in solution. Here are some additional thoughts on the zeta potential result: the negative zeta potential of the Ag@SeO2 bmNPs is likely due to the presence of functional groups on the surface of the nanoparticles, such as hydroxyl groups.53 These functional groups can dissociate in water, resulting in a negatively charged surface. The negative zeta potential of the Ag@SeO2 bmNPs can help to prevent the nanoparticles from aggregating.54 This is because nanoparticles with the same charge repel each other. The negative zeta potential can also help to improve their stability in solution. This is because the negative charge on the surface of the nanoparticles attracts water molecules, which creates a hydration layer around the nanoparticles. This hydration layer helps to protect the nanoparticles from aggregation and precipitation.
Dynamic light scattering (DLS) analysis revealed a polydisperse size distribution for Ag@SeO2 nanoparticles, indicating particles of various sizes. Two distinct peaks in the size distribution were observed: peak 1: smaller particles with an average size of 112.9 nm (SD 10 nm), likely representing individual Ag nanoparticles or small Ag@SeO2 composites. Peak 2: larger particles with an average size of 318.3 nm (SD 38.2 nm), likely representing larger Ag@SeO2 composite nanoparticles or even aggregates. The presence of two distinct populations and a broad size distribution (indicated by a PI value of 1.243) is further supported by the size distribution plot diameter. Analyzing the size distribution can help us optimize the synthesis process to achieve desired nanoparticle sizes for specific applications. By studying the factors influencing the distribution (e.g., precursor concentrations, reaction time, temperature), we can tailor the synthesis to obtain nanoparticles with optimal antibacterial or antioxidant properties.
The percent polydispersity of the Ag@SeO2 bmNPs is approximately 25%. This is relatively high, indicating that the Ag@SeO2 bmNPs are polydisperse. The interparticle forces between nanoparticles are dominated by van der Waals forces.55 These forces are attractive and can lead to the aggregation of the bmNPs. The surface chemistry of Ag@SeO2 bmNPs can also affect their aggregation. Hydrophilic bmNPs are less likely to aggregate than hydrophobic bmNPs. At higher concentrations, Ag@SeO2 bmNPs are more likely to collide and aggregate. In addition, a higher temperature can increase the kinetic energy of the nanoparticles, which can lead to an increased aggregation rate.56 The nanoparticles that are too small may be difficult to synthesize and characterize, while nanoparticles that are too large may not be able to penetrate cells or tissues. Crystalline nanoparticles have a well-defined structure, which gives them specific properties that can be exploited for different applications. For instance, crystalline nanoparticles may be more stable and durable than non-crystalline nanoparticles.
| D = K × λ/(β × cos(θ)) | (4) |
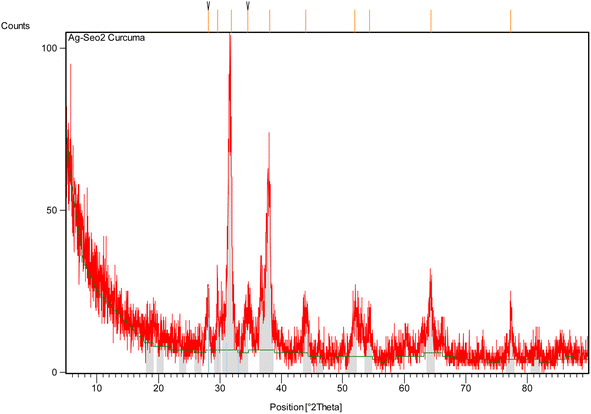
Using the Scherrer eqn (4), the average crystallite size of the Ag@SeO2 bmNPs was calculated to be approximately 20 nm. This is in good agreement with the size of the Ag@SeO2 bmNPs observed in TEM images. The FWHM values are generally small, indicating that the peaks are well-defined58 and that the Ag@SeO2 bmNPs have a high degree of crystallinity. The FWHM values increase slightly with decreasing d-spacing. This is expected, as smaller crystals tend to have broader peaks. The d-spacing values correspond to the crystal planes of Ag and SeO2, confirming the presence of both materials in the Ag@SeO2 bmNPs. The relative intensity values indicate that the (111) and (200) crystal planes of Ag are the most dominant in the Ag@SeO2 bmNPs. The relative intensity values of the Ag peaks are generally higher than those of the SeO2 peaks. This suggests that the Ag@SeO2 bmNPs have a higher surface area of Ag than SeO2. The presence of multiple peaks in the XRD pattern indicates that the nanoparticles are polycrystalline, meaning that they are composed of many small crystals. The data suggests that the nanoparticles are well-crystalline and have a core–shell structure with an Ag core and a SeO2 shell. The Ag@SeO2 bmNPs also have a high surface area of Ag. Mainly, the XRD analysis confirms the crystallinity, core–shell structure, and average size of the Ag@SeO2 bmNPs. This information can be used to optimize the biosynthesis process and to develop new applications for Ag@SeO2 bmNPs.
Table 2 summarizes the particles' size of Ag@SeO2 bmNPs calculated from XRD data using the Scherrer eqn (4). Peaks with similar d-spacings seem to have closer particle size estimations. For example, peaks 1 and 3 (both around 2.8 Å) have similar sizes (both around 25.4 nm), while peaks 5, 6, and 8 (around 2.0–2.3 Å) also have comparable sizes (around 14.6 nm).
| Peak number | Peak position (°2θ) | FWHM (°2θ) | Miller indices | d-Spacing (Å) | Calculated particle size (nm) |
|---|---|---|---|---|---|
| 1 | 28.0653 | 0.3149 | (111) | 3.17945 | 25.415 |
| 2 | 29.6122 | 0.2362 | (021) | 3.01679 | 30.568 |
| 3 | 31.8716 | 0.3149 | (121) | 2.80790 | 25.415 |
| 4 | 34.4904 | 0.9446 | (031) | 2.60046 | 8.554 |
| 5 | 38.0638 | 0.6298 | (131) | 2.36415 | 14.562 |
| 6 | 44.0496 | 0.6298 | (221) | 2.05579 | 14.562 |
| 7 | 51.9532 | 0.7872 | (230) | 1.76011 | 11.505 |
| 8 | 54.3324 | 0.6298 | (132) | 1.68853 | 14.562 |
| 9 | 64.3039 | 0.3936 | (321) | 1.44868 | 21.569 |
| 10 | 77.3277 | 0.3840 | (400) | 1.23297 | 22.143 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
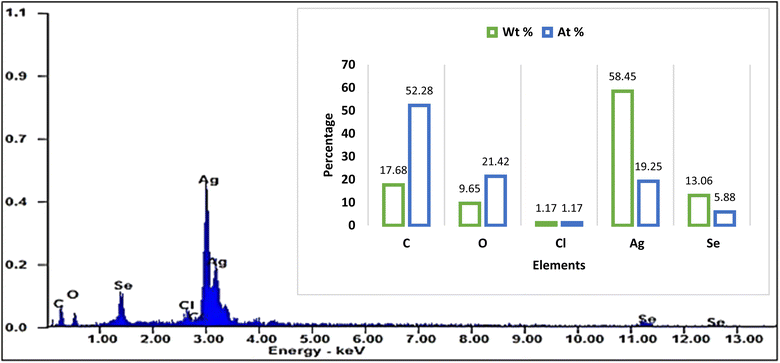
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which is consistent with the stoichiometry of Ag@SeO2 bmNPs. Particularly, the occurrence of carbon and oxygen suggests that the Ag@SeO2 bmNPs are coated with a layer of organic matter. This organic layer may help to stabilize the Ag@SeO2 bmNPs and prevent them from agglomerating.59 The high atomic ratio of Ag to Se indicates that the Ag@SeO2 bmNPs are composed of a single layer of Ag NPs on a SeO2 core. This core–shell structure is expected to give the Ag@SeO2 bmNPs unique properties, such as enhanced catalytic activity and antimicrobial activity.
1, which is consistent with the stoichiometry of Ag@SeO2 bmNPs. Particularly, the occurrence of carbon and oxygen suggests that the Ag@SeO2 bmNPs are coated with a layer of organic matter. This organic layer may help to stabilize the Ag@SeO2 bmNPs and prevent them from agglomerating.59 The high atomic ratio of Ag to Se indicates that the Ag@SeO2 bmNPs are composed of a single layer of Ag NPs on a SeO2 core. This core–shell structure is expected to give the Ag@SeO2 bmNPs unique properties, such as enhanced catalytic activity and antimicrobial activity.
Silver nanoparticles might be adsorbed onto the surface of SeO2 nanoparticles. The nanoparticles could be individual entities that have clumped together, appearing smooth on the surface. One element might be concentrated on the outer surface, even if not in a perfect shell-like structure. The selenium atoms of the Ag@SeO2 bmNPs are likely arranged in a hexagonal closed-packed (hcp) crystal structure.60,61 This is the most stable crystal structure for selenium. Furthermore, the Ag@SeO2 bmNPs are likely polycrystalline, meaning that they are composed of many small crystals. The size of the crystals is likely in the range of 10–20 nm.
3.2. The mechanism of the formation of Ag@SeO2 bmNPs
The formation of Ag@SeO2 bmNPs core–shell nanoparticles involves a multi-step process that encompasses both reduction and oxidation reactions.62,63 The first step is the reduction of AgNO3 to Ag-NPs by turmeric extract. Turmeric extract, rich in polyphenolic compounds like curcumin, acts as a natural reducing agent capable of converting silver ions (Ag+) to silver nanoparticles (Ag-NPs) in silver nitrate (AgNO3) solution. The hydroxyl groups and methoxy groups present in curcumin can donate electrons to Ag+, leading to the formation of Ag atoms and the stabilization of Ag-NPs by curcumin molecules. This reduction reaction can be represented as:| 2AgNO3 + C14H16O6(curcumin) → 2Ag + 2HNO3 + C14H16O6(oxidized curcumin) |
The second step is the formation of Ag@SeO2 bmNPs. In which, selenium dioxide (SeO2) dissociates in water to form selenite ions (SeO32−). Ag-NPs (formed in the first step) interact with SeO32− where Ag-NPs acts as a reducing agent towards SeO32− ions. The SeO32− ions are reduced to selenide ions (Se2−) and adsorbed onto the surface of the Ag-NPs. This reduction reaction can be represented as:
| Ag + SeO32− → Ag@SeO2 bmNP + Se2− |
During the formation of Ag@SeO2 bmNP, the adsorbed Se2− ions undergo oxidation in the presence of oxygen (O2) to form a thin layer of SeO2 on the surface of the Ag-NPs, resulting in the formation of Ag@SeO2 bmNPs core–shell nanoparticles. This oxidation reaction can be represented as:
| Se2− + O2 → SeO2 |
The overall process can be summarized by the following simplified reactions:
| 2AgNO3 + C14H16O6(curcumin) → 2Ag + 2HNO3 + C14H16O6(oxidized curcumin) |
| Ag + SeO32− → Ag@SeO2 bmNP + Se2− |
| Se2− + O2 → SeO2 |
Factors influencing the formation and properties of Ag@SeO2 bmNPs core–shell nanoparticles include the concentration of turmeric extract, AgNO3, and SeO2; temperature and reaction time; pH of the solution, and the presence of stabilizing or capping agents (e.g., turmeric extract). The resulting Ag@SeO2 bmNPs core–shell nanoparticles exhibit unique properties due to the synergistic combination of AgNPs and SeO2, making them promising materials for various applications. The next investigation was to optimize the biosynthesis process of Ag@SeO2 bmNPs.
3.3. Modeling Ag@SeO2 bmNPs biosynthesis by BBD
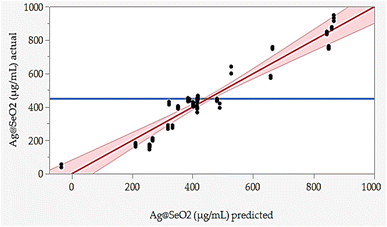
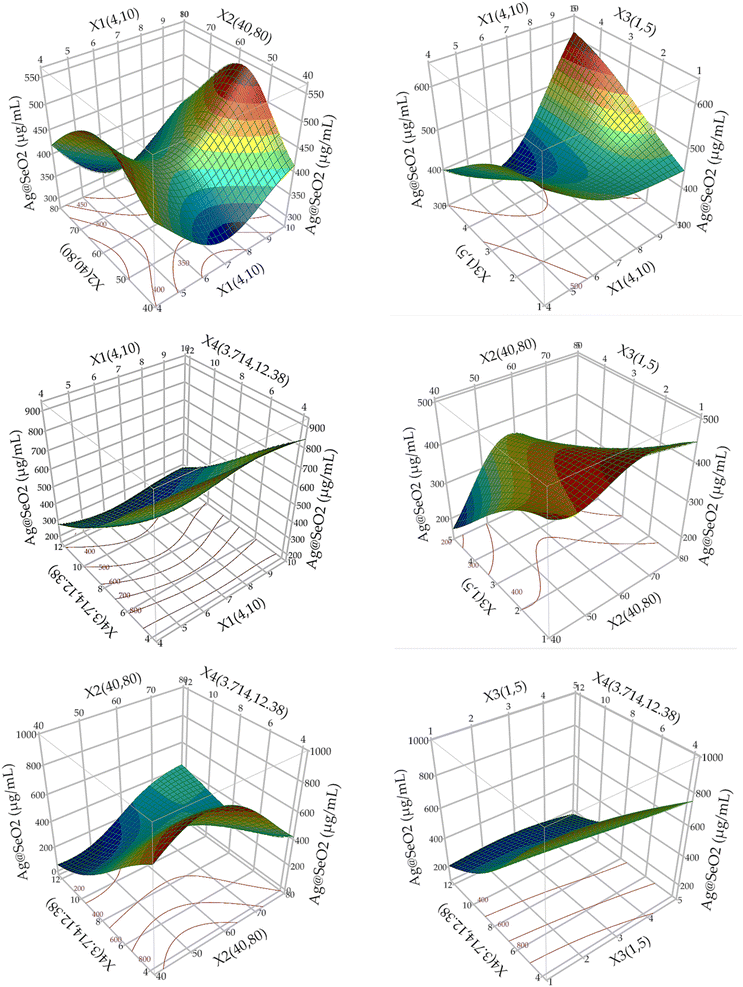
The responded data in Table 1 present the results of a BBD experiment for the Ag@SeO2 bmNPs synthesis using turmeric extract. The data demonstrate the influence of four critical factors i.e., pH (X1), bioconversion temperature (X2), bioconversion time (X3), and turmeric concentration (X4) on the Ag@SeO2 bmNPs yield. The relative agreement between the predicted and actual values of BBD validates the model's effectiveness.Furthermore, the error analysis shows the difference between the actual and predicted values, indicating how well the model fits the data. The model predictions generally agree with the actual values in most points, as evidenced by the overall small error values. However, some runs show obvious discrepancies between predicted and actual values, such as runs 24, 42, 43, 48, 72, and 78. This suggests that the model may need further refinement. However, the linear relationship between the predicted and actual points was plotted (Fig. 7).
The BBD model displayed points that aligned with the prediction line. Nevertheless, linear regression demonstrated that several points are notably located away from the prediction line. Overall, the data suggests an obvious relationship between the factors and Ag@SeO2 bmNPs production, as evidenced by the variability in the actual values. These findings suggest the potential of turmeric extract as a natural and efficient reducing agent for Ag@SeO2 bmNP synthesis, paving the way for further research into the underlying mechanisms and applications of these biogenic nanoparticles (Fig. 7).
3.4. Evaluation of the BBD model
The ANOVA data (Table 3) provides valuable insights into the main and interactive effects of various factors on Ag@SeO2 bmNPs biosynthesis using turmeric extract. ANOVA confirms the effectiveness of the BBD in capturing the significant factors influencing Ag@SeO2 bmNP biosynthesis using turmeric extract. The model exhibits a good adjusted-R2 value of 0.9075, indicating that it accounts for 90.75% of the data's variability. Additionally, the predicted-R2 value of 0.8829 suggests that the model may have a good predictive ability.| Source | Freedom degree | Sum of squares | Mean square | F-Ratio | p-Value |
|---|---|---|---|---|---|
| a X1; pH, X2; bioconversion temperature (°C), X3; bioconversion time (h), X4; concentration of turmeric extract (mg mL−1).b Significant value. | |||||
| Model | 14 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682 682![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 206 206 |
263![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 015 015 |
57.07 | <0.001b |
| Linear | 4 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 638 638![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 530 |
659![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 633 633 |
143.13 | <0.001b |
| X1 | 1 | 5 | 5 | 0.00 | 0.973 |
| X2 | 1 | 40 | 40 | 0.01 | 0.926 |
| X3 | 1 | 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 967 967 |
150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 967 967 |
32.76 | <0.001b |
| X4 | 1 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487 487![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 518 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487 487![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 518 518 |
539.76 | <0.001b |
| Square | 4 | 548![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 469 469 |
137![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 117 117 |
29.75 | <0.001b |
| X1 × X1 | 1 | 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 483 483 |
123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 483 483 |
26.79 | <0.001b |
| X2 × X2 | 1 | 141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 732 732 |
141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 732 732 |
30.75 | <0.001b |
| X3 × X3 | 1 | 30 | 30 | 0.01 | 0.936 |
| X4 × X4 | 1 | 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 870 870 |
117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 870 870 |
25.58 | <0.001b |
| 2-Way interaction | 6 | 495![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 207 207 |
82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 534 534 |
17.91 | <0.001b |
| X1 × X2 | 1 | 598 | 598 | 0.13 | 0.720 |
| X1 × X3 | 1 | 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
20.27 | <0.001b |
| X1 × X4 | 1 | 750 | 750 | 0.16 | 0.688 |
| X2 × X3 | 1 | 19 | 19 | 0.00 | 0.948 |
| X2 × X4 | 1 | 384![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 392 392 |
384![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 392 392 |
83.41 | <0.001b |
| X3 × X4 | 1 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 047 047 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 047 047 |
3.48 | 0.066 |
| Error | 66 | 304![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 164 164 |
4609 | ||
| Lack-of-fit | 10 | 291![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185 185 |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 118 118 |
125.63 | <0.001b |
| Pure error | 56 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 979 979 |
232 | ||
| Total | 80 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 986 986![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 370 370 |
|||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Model evaluation statistics | |||||
| Determination coefficient (R2) | 0.9237 | ||||
| Adjusted-R2 | 0.9075 | ||||
| Predicted-R2 | 0.8829 | ||||
One of the important model goodness relies on high positive R2 and adjusted-R2 values that are ≥0.75 and relatively close.64,65 The current model boasts high values (R2 = 0.9237, adjusted-R2 = 0.9075), suggesting strong agreement between predicted and experimental results. Increasing R2 and adjusted-R2 values generally signify an improved model fit and more accurate relationships between tested variables and the response. However, a high R2 alone isn't a guarantee of model quality. Only when accompanied by a similarly high adjusted-R2 and an insignificant lack-of-fit can truly infer a robust regression model.66
Regarding the tested variables, two (X3, and X4) of the four investigated factors exhibit statistically significant individual effects on Ag@SeO2 bmNP production. Notably, turmeric concentration has the most significant impact, with the highest F value and a low p-value of less than 0.001, highlighting its crucial role in the biosynthesis process. The two-way interaction between bioconversion temperature and turmeric concentration stands out as statistically significant (p-value < 0.001), indicating a synergistic effect between these two factors. Similarly, the interaction between pH and bioconversion time shows a significant influence (p-value < 0.001). The square effect of bioconversion time was the only insignificant factor.
To decipher the influence of individual variables and their interactions on Ag@SeO2 bmNP biosynthesis, statistically significant coefficients are key. Typically, an alpha level of ≤0.05 is set to assess the significance of model terms.67 At this threshold, a significant coefficient indicates a less than 5% chance of wrongly concluding a link between a variable and the response when none exists.66,68 Unfortunately, our model exhibits a significant lack-of-fit (p-value < 0.001), hinting at unaccounted-for factors or potential experimental errors. This highlights the need for further optimization and exploration of additional variables to refine the model's accuracy and predictive power.
The ANOVA results for the quadratic model (Table 3), investigated the influence of experimental variables on Ag@SeO2 bmNP biosynthesis. The highly significant model (p < 0.001) and high R2 values (R2 = 0.9237, adjusted-R2 = 0.9075) indicate a strong correlation between the variables and the response. However, the significant lack-of-fit (p < 0.001) necessitates potential model improvement.
Despite the statistically significant findings of the BBD in capturing the main and interactive effects of factors influencing Ag@SeO2 bmNP biosynthesis, the significant lack-of-fit test suggests limits its ability to fully model the complex relationships involved. The lack-of-fit test reveals a significant discrepancy between the predicted values generated by the Box–Behnken model and the actually observed Ag@SeO2 bmNP yield (p-value < 0.001). This suggests that the model, while informative, doesn't perfectly capture the complex relationships between the investigated factors and the response variable (Ag@SeO2 bmNP yield). This necessitates the exploration of alternative modeling approaches to improve the accuracy and predictive capabilities of the model. While the current model provides valuable information about the main and interactive effects of the investigated factors, the lack-of-fit highlights its limitations. So, another modeling approach is necessary to enhance the predictive capabilities and reliability.
3.5. Modeling Ag@SeO2 bmNP by ANN
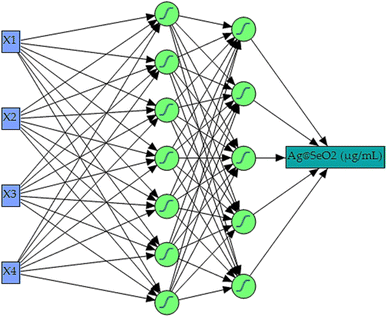
Maximizing Ag@SeO2 bmNP bioproduction demanded a cutting-edge AI-powered modeling approach. Building upon Box–Behnken design data, a fully connected, multilayer feed-forward ANN was constructed as the predictive core of the neural network platform. Extensive trial-and-error optimization, involving numerous iterations of 5000 training epochs each, yielded the ideal ANN configuration. This optimal model, featuring a squared learning rate of 0.1, one input layer, one output layer, and two hidden layers, leveraged a 0.3333 holdback propagation for data partitioning. This split the data into two distinct sets: a 54-run training set for error reduction and weight establishment, and a 27-run validation set for fine-tuning model selection.Within the hidden layers, all nodes utilized a shared NTanH activation function, known as, for optimized signal processing. This configuration, alongside the determined optimal network topology of 3–5–7–1 neurons (Fig. 8), facilitated superior model performance. The four input neurons directly corresponded to the investigated independent factors (pH, bioconversion temperature, time, and turmeric extract concentration). Meanwhile, the single output neuron served as the sole predictor for Ag@SeO2 bmNP biosynthesis. Notably, the NTanH activation function effectively supported the performance of both hidden layers, contributing to the overall model effectiveness.
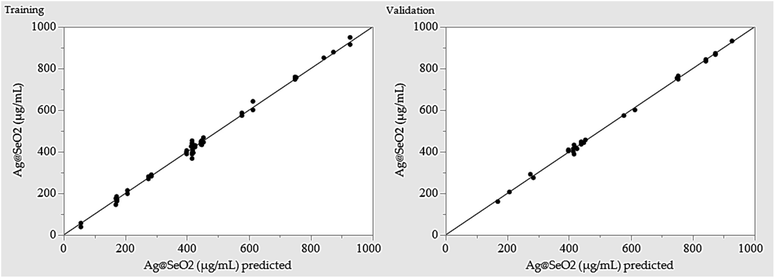
Training prioritized maximizing the R2, ensuring the model closely mirrors reality. Once trained, the network's predictive power was rigorously evaluated by assessing its ability to generate outputs closely resembling, or even identical to the actual Ag@SeO2 bmNP values. Table 1 showcases the predicted values generated by the ANN model. Comparing them to the experimental data reveals a remarkable agreement, with the ANN outperforming the BBD model by exhibiting lower residual gaps.
The rise of artificial intelligence offers powerful tools for scientific research. Among these, ANNs stand out for their versatility and predictive prowess. Their unique architecture, employing flexible functions for input–output mapping,69 makes them particularly suitable for our investigation of Ag@SeO2 bmNP biosynthesis using turmeric extract. Unlike traditional modeling techniques, ANNs are not limited by linear relationships and can excel with the right architecture.67 However, unlike response surface designs, the black box nature of ANN limits the delving into complex systems of the input–output relationships.67,70,71 Nevertheless, the apparent mutual relationship is not crucial for our study, as the metal relationships of biological processes behind Ag@SeO2 bmNP biosynthesis were not our aim. Finally, ANNs can learn from diverse data types by tailoring their internal structure, i.e., nodes and hidden layers.68 This flexibility empowers us to build an efficient model even with potentially limited or complex data.
3.6. Evaluation of the ANN model
The 3D plots showcased diverse patterns, this reflects ANN's proficiency in deciphering complex, nonlinear associations between inputs and outputs, regardless of their apparent relationship. Hidden layers facilitate this process, enabling indirect pathways between inputs and outputs, unlike conventional models with direct paths.73 This makes ANNs exceptional predictors even when input–output relationships are unclear or irrelevant.66,70,71
Sarkar et al.74 pointed out that the biosynthesis of silver nanoparticles occurred in pH ranging from 5.5 to 8.0, while the stability of Ag NPs was synthesized at an alkaline pH, and adequate stability was noted at pH 7. Additionally, the optimization process of Ag NPs synthesis was significantly affected by the concentration of AgNO3, dose of extract, and bioconversion time, whereas, the temperature of NPs synthesis did not have a significant effect. Liaqat et al.75 optimized, they found that the optimum reaction time of Ag NPs green synthesis was 60 min, the optimum temperature was 75 °C, and the optimum concentration of AgNO3 was 1 mM. Another study reported that the optimization process of biosynthesized AgNPs was conducted under pH 7, reaction temperature of 25 °C, 1 mM AgNO3 concentration, and 15–20 g of wet biomass of Penicillium sp.76 As well as, the optimized selenium nanoparticles were achieved at pH 7, and 32 °C by Streptomyces sp.77 Birla et al.78 reported the biosynthesis and optimization of silver nanoparticles at a pH range from 9 to 11 and a temperature range from 40–60 °C. As well as, they found that there are correlations between the yield of nanoparticles and the volume of filtrate, biomass quantity, and salt concentration.
3.7. Model performance
Initial evaluation revealed the ANN model's robust predictive capabilities, evident in minimal residuals and accurate fit to experimental data. Performance metrics calculated during training, validation, and overall stages (Table 4) confirmed this, with R2 values exceeding 0.99 and low error values (root average square error and mean absolute deviation). This demonstrates the model's high confidence and accuracy in predicting Ag@SeO2 bmNP biosynthesis.| Measure | Training | Validation | Overall model |
|---|---|---|---|
| R2 | 0.9953 | 0.9980 | 0.9965 |
| Root average square error | 14.687 | 9.549 | 13.198 |
| Mean absolute deviation | 10.876 | 7.401 | 9.718 |
| Sum frequency | 54 | 27 | 81 |
3.8. Optimum conditions and model validation
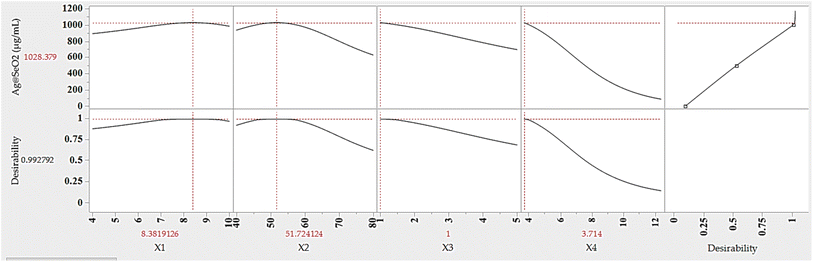
The ANN model was used to predict the optimum levels of the four tested factors that maximize Ag@SeO2 bmNP biosynthesis conditions (Fig. 12). The calculated values were found to be 9.83 pH, 51.7 °C bioconversion temperature, 1.0 h bioconversion time, and 3.71 mg mL−1 of turmeric extract. Under such conditions, the response of Ag@SeO2 bmNP biosynthesis was calculated to be 1028.38 μg mL−1, with a desirability value of 0.9928. Subsequently, the estimated optimum conditions for the 4 variables were verified through laboratory experimentation. The validation test revealed high reliability with the actual Ag@SeO2 bmNP biosynthesis yield, being 1034.53 ± 17.38 μg mL−1, exhibiting high performance of the ANN model.To pinpoint the ideal conditions for maximizing Ag@SeO2 bmNP biosynthesis, the desirability function was used. These versatile tools act as guides, mathematically evaluating different scenarios and assigning scores ranging from 0 (undesirable) to 1 (highly desirable), this is considered a crucial step before model validation to set clear targets for prediction accuracy.28,64,79 The predicted values aligned remarkably with the experimental findings, affirming the desirability function's success in guiding us toward optimal conditions for biosynthesis.
Ruling the realm of complex systems, ANNs are the undisputed navigators, effortlessly unraveling riddles of nonlinearity and multicollinearity compared with the other models. With their intricate internal compass, they chart hidden pathways and uncover cryptic connections, revealing secrets that simpler models could never do. While other models might take a direct path to the target, ANNs embrace a more intricate journey, weaving through hidden layers and iterations to unlock hidden patterns.28,29 This meticulous approach yields impressive predictive accuracy, but it also comes with trade-offs. The process demands time for modeling, and the intricate connections can make it challenging to pinpoint the exact role of individual factors within the model.71,80 This complexity can sometimes limit the ability to simplify the model or isolate individual factors for further exploration.
3.9. Phytochemical analyses
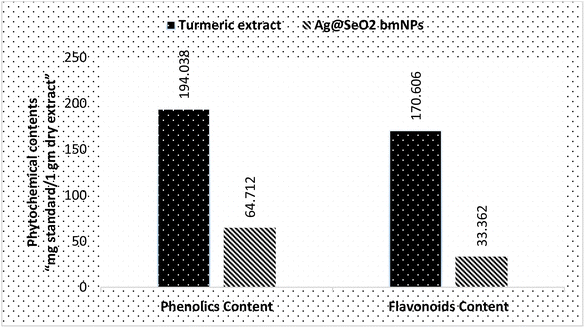
The phytochemical analyses (Fig. 13 and Table S4†) revealed that turmeric extract has a significantly higher phenolics content (194.038 ± 2.8 mg g−1) compared to Ag@SeO2 bmNPs (64.712 ± 1.3 mg g−1). This suggests that turmeric extract is a richer source of phenolic compounds, which are known to have antioxidant and other health-promoting properties. Similar to phenolics content, turmeric extract also has a higher flavonoid content (170.606 ± 0.59 mg g−1) compared to Ag@SeO2 bmNPs (33.362 ± 0.74 mg g−1). The high content of phenolics and flavonoids in the turmeric extract indicates the potential for strong antioxidant activity. Based on the literature reports, turmeric extract is a rich source of phenolic content such as curcuminoids (e.g., curcumin, dimethoxy-curcumin, and bis-dimethoxy-curcumin),81,82 which are the most abundant phenolic compounds in turmeric, responsible for its characteristic yellow color. In addition, turmeric extract includes other types of phenolic contents such as ferulic acid benefits cardiovascular and skin health, vanillin “contributes to the pleasant flavor of turmeric”, and p-coumaric acid “phenolic acid”. Turmeric extract on the other side also is a rich source of flavonoid contents which contributes to its antioxidant activity such as quercetin, kaempferol, gallic acid, and naringenin cholesterol-lowering effects.83,84Phytochemicals e.g. phenolic compounds like curcumin readily donate electrons to metal ions, triggering their reduction to MNPs.85 These electron-rich molecules act as mild and biocompatible alternatives to harsh chemical-reducing agents used in conventional synthesis.86 Phytochemicals also prevent the newly formed MNPs from aggregating and growing uncontrollably. Phytochemicals often possess functional groups like hydroxyl or amine groups that bind to the MNP surface, providing steric hindrance and electrostatic repulsion.87 This capping ability ensures size control and stability, crucial for desired MNP properties. Phytochemicals influence the size, shape, and morphology of the resulting MNPs.88 Specific interactions between phytochemicals and metal ions can direct the nucleation and growth process, leading to MNPs with tailored properties for specific applications.
3.10. Antioxidant activity
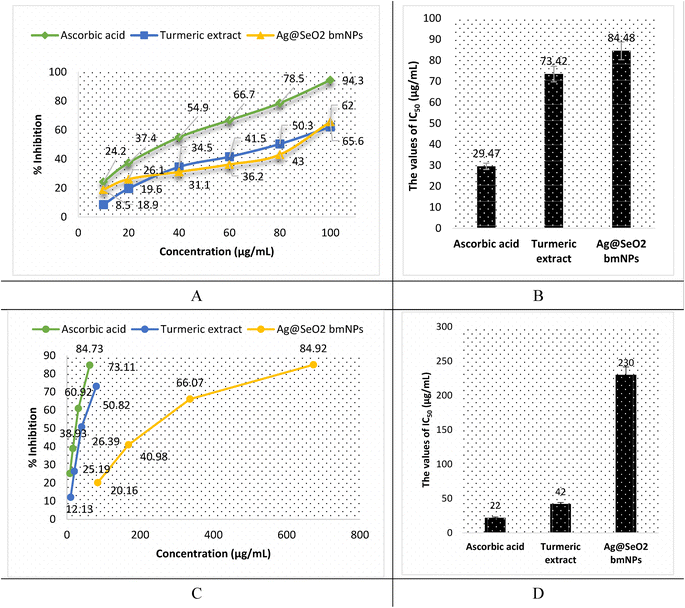
The antioxidant activity of turmeric extract, Ag mNPs, and Ag@SeO2 bmNPs was assessed by ABTS assay. The results (Fig. 14A, B and Table S5†) showed that turmeric extract and Ag@SeO2 bmNPs have antioxidant activity, but turmeric extract is the most potent antioxidant. The IC50 values for turmeric extract, Ag@SeO2 bmNPs, and ascorbic acid are 29.47, 73.42, and 84.48 μg mL−1, respectively. This means that turmeric extract can inhibit the ABTS radical by 50% at a concentration that is lower than the concentration of Ag@SeO2 bmNPs, or ascorbic acid. The antioxidant activity of turmeric extract is likely due to the presence of curcuminoids, which are polyphenols with strong antioxidant properties. Curcuminoids have been shown to scavenge free radicals, chelate metal ions, and inhibit the formation of reactive oxygen species (ROS). The antioxidant activity of Ag@SeO2 bmNPs is likely due to the presence of silver nanoparticles, which have been shown to scavenge free radicals and inhibit the formation of ROS. Ag@SeO2 bmNPs may also have enhanced antioxidant activity due to the presence of selenium, which is an essential nutrient with antioxidant properties.Briefly, the results of the ABTS assay show that turmeric extract, and Ag@SeO2 bmNPs all have antioxidant activity, but turmeric extract is the most potent antioxidant. This suggests that turmeric extract may be a useful dietary supplement or ingredient in functional foods for the prevention of oxidative stress and related diseases. In addition, the antioxidant activity of turmeric extract, and Ag@SeO2 bmNPs increases with increasing concentration. This is expected, as antioxidants typically scavenge free radicals in a dose-dependent manner. The antioxidant activity of turmeric extract is comparable to that of ascorbic acid, which is a known antioxidant. This suggests that turmeric extract may be an effective alternative to ascorbic acid for preventing oxidative stress and related diseases. The results of this study are promising and suggest that turmeric extract and Ag@SeO2 bmNPs may have the potential as antioxidants for the prevention and treatment of oxidative stress-related diseases.
The antioxidant activity by DPPH assay is shown in Fig. 14C, D and Table S6.† Turmeric extract demonstrates potent antioxidant activity, suggesting strong free radical scavenging potential. The IC50 value of 42 μg mL−1 of turmeric extract indicates that it is comparable and effective to ascorbic acid (IC50 = 22 μg mL−1) in this specific assay. In addition, turmeric extract shows strong dose-dependent antioxidant activity, scavenging up to 73.11% of DPPH radicals at the lowest concentration (80 μg mL−1). However, Ag@SeO2 bmNPs revealed lower antioxidant activity than the turmeric extract, as reflected by the higher IC50 value (230 μg mL−1). Ag@SeO2 bmNPs displayed weaker antioxidant activity compared to turmeric extract at all concentrations and showed a dose-dependent increase in DPPH scavenging but reached only 84.92% at the highest concentration (673 μg mL−1). Both ABTS and DPPH assays show that turmeric extract exhibits strong antioxidant activity compared to Ag@SeO2 bmNPs.
The mechanism of action was illustrated as the turmeric extract contains curcuminoids, which are polyphenols with strong antioxidant properties.89 Curcuminoids can scavenge free radicals, such as superoxide radicals, hydroxyl radicals, and peroxyl radicals, by donating hydrogen atoms or electrons. Turmeric extract contains curcuminoids, which can chelate metal ions and prevent them from catalyzing free radical formation.90 ROS, such as superoxide radicals and hydrogen peroxide, are produced during normal cellular metabolism. However, excessive ROS production can lead to oxidative stress and cell damage. Curcuminoids can inhibit the formation of ROS by inhibiting enzymes that produce ROS, such as NADPH oxidase and xanthine oxidase.91 There are a few reasons why nanoparticles may be less potent antioxidants than turmeric extract: the size and shape of nanoparticles can affect their antioxidant activity. Smaller nanoparticles have a larger surface area, which allows them to interact with and scavenge free radicals more efficiently.92 However, nanoparticles can also agglomerate, which can reduce their surface area and antioxidant activity.
Turmeric extract contains a variety of curcuminoids, which have different sizes and shapes. This variety of sizes and shapes may allow curcuminoids to interact with and scavenge a wider range of free radicals. The surface chemistry of nanoparticles can also affect their antioxidant activity. Nanoparticles with a positively charged surface tend to be more potent antioxidants than nanoparticles with a negatively charged surface.93 This is because positively charged nanoparticles can attract and scavenge free radicals more efficiently. Nanoparticles can interact with biological systems in complex ways.94 Some nanoparticles can be toxic to cells, while others can be taken up by cells and metabolized. The biocompatibility of nanoparticles can affect their antioxidant activity. For example, if nanoparticles are toxic to cells, they may damage cells and generate oxidative stress.95 This would reduce the antioxidant activity of the nanoparticles. Turmeric extract is generally well-tolerated by humans and has a low toxicity profile. This makes it a more biocompatible antioxidant than nanoparticles.
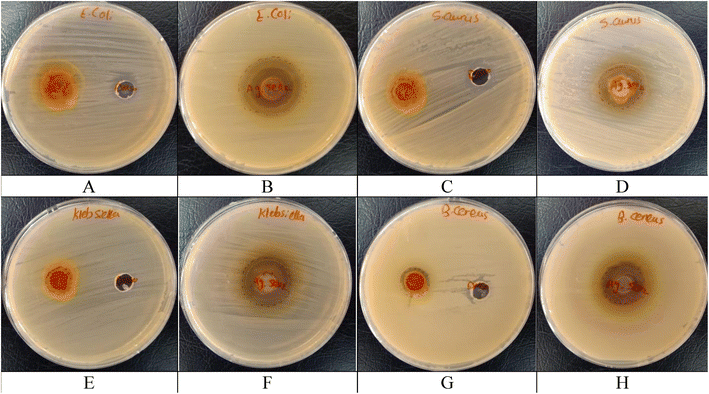
3.11. Antibacterial activity
| Samples | Inhibition zone diameters (mm) | |||
|---|---|---|---|---|
| E. coli (ATCC 10536) | S. aureus (ATCC 6538) | K. pneumoniae (ATCC 10031) | B. cereus (EMCC number 1080) | |
| Turmeric extract | −ve | 13 ± 1.61 | 13 ± 1.19 | 14 ± 2.06 |
| Ag@SeO2 bmNPs | 32 ± 1.25 | 22 ± 0.79 | 25 ± 0.82 | 22 ± 1.03 |
| Control (DMSO) | −ve | −ve | −ve | −ve |
| Cefotaxime | 23 ± 0.95 | 11 ± 2.50 | 25 ± 1.74 | 20 ± 1.96 |
The moderate and broad-spectrum activity of turmeric extract suggests a multi-target or non-specific mechanism of action. The possible mechanism of action could be suggested as curcumin, a major component of turmeric, is known to interact with bacterial membranes, potentially causing leakage of essential components and cell death.96 Curcumin and other bioactive compounds in the extract might inhibit key enzymes involved in bacterial metabolism or protein synthesis, hindering their growth and survival.97 Curcumin exhibits antioxidant properties that could scavenge free radicals generated by bacteria, causing oxidative stress and damage.98 Turmeric extract could potentially interfere with bacterial communication pathways, disrupting their ability to coordinate defense mechanisms or biofilm formation.99
The potent and broad-spectrum activity of Ag@SeO2 bmNPs points towards a more direct and efficient mechanism of action, likely involving silver ions released from the nanoparticles. The mechanism of action of Ag@SeO2 bmNPs was commended as follows: silver ions bind to thiol groups on bacterial proteins and membranes, causing disruption and leakage of essential components.100 This explains the potent and broad-spectrum activity of Ag@SeO2 bmNPs. Silver ions can induce the production of ROS within bacteria, causing oxidative stress and damage to DNA, proteins, and other cellular structures.101 Silver ions can bind to bacterial DNA, inhibiting its replication and preventing cell division.102
El-deeb et al.77 found that Ag NPs have potential activity towards E. coli ranging 13–20 mm inhibition zone under different pH values. Kim et al.103 showed a similar effect of Ag NPs against E. coli and S. aureus. Singh et al.76 found the antibacterial activity of Ag NPs against MDR E. coli and S. aureus being 17 and 16 mm inhibition zones, respectively. Ninganagouda et al.104 reported that the Ag NPs showed potentiality against E. coli. The SeO2 coating on the nanoparticles might enhance the antibacterial activity by generating additional ROS or acting as a slow-release reservoir for silver ions.105 Frequently, the results presented in Table 4 warrant further investigation of both turmeric extract and Ag@SeO2 bmNPs as potential alternatives or supplements to Cefotaxime. Their diverse activities and promising initial results encourage continued research to elucidate their mechanisms of action, optimize their effectiveness, and explore their potential for clinical applications.
| Bacteria strain | MIC (μg mL−1) | O.D. 600 at MIC |
|---|---|---|
| B. cereus | 331.25 | 0.007 |
| K. pneumoniae | 331.25 | 0.001 |
| E. coli | 165.625 | 0.001 |
| S. aureus | 165.625 | 0.001 |
4. Conclusion
Phytochemicals in turmeric extract perform a crucial role in reducing SeO2 into Ag@SeO2 bmNPs. Their electron-donating capabilities drive the reduction, while their functional groups contribute to nanoparticle stabilization and shape control. This green and sustainable approach overlays the way for successful biosynthesis of Ag@SeO2 bmNPs, with various applications and desirable characteristics. Ag@SeO2 bmNPs are characterized by their negative surface charge, spherical morphology, crystalline nature, core–shell structure, and consistent elemental composition. Combining BBD and ANN modeling effectively optimizes Ag@SeO2 bmNP biosynthesis using turmeric extract. ANN demonstrated superior predictive accuracy and identified optimal conditions for maximizing yield. However, its black-box nature necessitates further investigation into individual factor effects and exploration of alternative modeling approaches for deeper mechanistic understanding and process refinement. Turmeric extract is a rich source of bioactive compounds with potent antioxidant and antibacterial properties. Bio-synthesized Ag@SeO2 bmNPs show promising antioxidants and their potential activity against various bacterial strains. Ultimately, further investigation is needed to elucidate the exact mechanisms of action and optimize the effectiveness of both turmeric extract and bmNPs for potential applications in food preservation, medicine, and other fields.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work is supported by Researchers Supporting Project number (RSP2024R114), King Saud University, Riyadh, Saudi Arabia.References
- D. Sharma, S. Kanchi and K. Bisetty, Arabian J. Chem., 2019, 12(8), 3576–3600 CrossRef CAS.
- V. Soni, P. Raizada, P. Singh, H. N. Cuong, S. Rangabhashiyam, A. Saini, R. V. Saini, Q. Van Le, A. K. Nadda, T. T. Le and V. H. Nguyen, Environ. Res., 2021, 202, 111622 CrossRef CAS PubMed.
- A. Amalraj, A. Pius, S. Gopi and S. Gopi, J. Tradit. Complementary Med., 2017, 7(2), 205–233 CrossRef PubMed.
- I. Chattopadhyay, K. Biswas, U. Bandyopadhyay and R. K. Banerjee, Curr. Sci., 2004, 44–53 CAS.
- J. Sharifi-Rad, Y. E. Rayess, A. A. Rizk, C. Sadaka, R. Zgheib, W. Zam, S. Sestito, S. Rapposelli, K. Neffe-Skocińska, D. Zielińska and B. Salehi, Front. Pharmacol, 2020, 11, 1021 CrossRef CAS PubMed.
- R. K. Verma, P. Kumari, R. K. Maurya, V. Kumar, R. B. Verma and R. K. Singh, Int. J. Chem. Stud., 2018, 6(4), 1354–1357 Search PubMed.
- S. Jaiswal and P. Mishra, Med. Microbiol. Immunol., 2018, 207, 39–53 CrossRef CAS PubMed.
- S. Chhabria and K. Desai, Encycl. Nanosci. Nanotechnol., 2016, 20, 1–32 Search PubMed.
- M. Y. Ali, W. K. Abad, H. M. Roomy and A. N. Abd, NanoWorld J., 2023, 9(3), 89–93, DOI:10.17756/nwj.2023-121.
- R. D. Sarkar and M. C. Kalita, Heliyon, 2022, 8(3), e09076 CrossRef CAS PubMed.
- D. Medina-Cruz, B. Saleh, A. Vernet-Crua, A. Nieto-Argüello, D. Lomelí-Marroquín, L. Y. Vélez-Escamilla, J. L. Cholula-Díaz, J. M. García-Martín and T. Webster, Racing for the Surface: Antimicrobial and Interface Tissue Engineering, Springer, Cham, 2020, pp. 397–434, DOI:10.1007/978-3-030-34471-9_16.
- I. Ali, C. Peng, I. Naz, M. A. Amjed, Magnetic Nanostructures: Environmental and Agricultural Applications, 2019, pp. 161–179 Search PubMed.
- D. Sári, A. Ferroudj, S. Dávid, H. El-Ramady, M. Abowaly, Z. F. Abdalla, H. Mansour, Y. Eid and J. Prokisch, Egypt. J. Soil Sci., 2024, 64(1), 1–17 Search PubMed.
- M. I. Din and A. Rani, Crit. Rev. Anal. Chem., 2018, 48(5), 406–415 CrossRef CAS PubMed.
- V. Soni, P. Raizada, P. Singh, H. N. Cuong, S. Rangabhashiyam, A. Saini, R. V. Saini, Q. Van Le, A. K. Nadda, T. T. Le and V. H. Nguyen, Environ. Res., 2021, 202, 111622 CrossRef CAS PubMed.
- M. Saravanan, H. Barabadi and H. Vahidi, Green nanotechnology: isolation of bioactive molecules and modified approach of biosynthesis, in Biogenic Nanoparticles for Cancer Theranostics, Elsevier, 2021, pp. 101–122 Search PubMed.
- B. R. Cuenya, Thin Solid Films, 2010, 518(12), 3127–3150 CrossRef CAS.
- R. Zhang, A. Khalizov, L. Wang, M. Hu and W. Xu, Chem. Rev., 2012, 112(3), 1957–2011 CrossRef CAS PubMed.
- S. Karthikeyan and B. Ramamoorthy, Appl. Surf. Sci., 2014, 307, 654–660 CrossRef CAS.
- R. Javed, M. Zia, S. Naz, S. O. Aisida, N. U. Ain and Q. Ao, J. Nanobiotechnol., 2020, 18, 1–15 CrossRef PubMed.
- B. Kowalczyk, K. J. Bishop, I. Lagzi, D. Wang, Y. Wei, S. Han and B. A. Grzybowski, Nat. Mater., 2012, 11(3), 227–232 CrossRef CAS PubMed.
- T. Østergård, R. L. Jensen and F. S. Mikkelsen, Energy Build., 2020, 208, 109628 CrossRef.
- A. Jankovic, G. Chaudhary and F. Goia, Energy Build., 2021, 250, 111298 CrossRef.
- H. Tao, T. Wu, M. Aldeghi, T. C. Wu, A. Aspuru-Guzik and E. Kumacheva, Nat. Rev. Mater., 2021, 6(8), 701–716 CrossRef.
- Y. Sun, Chem. Soc. Rev., 2013, 42(7), 2497–2511 RSC.
- S. S. Salem and A. Fouda, Biol. Trace Elem. Res., 2021, 199, 344–370 CrossRef CAS PubMed.
- X. Y. Yang, L. H. Chen, Y. Li, J. C. Rooke, C. Sanchez and B. L. Su, Chem. Soc. Rev., 2017, 46(2), 481–558 RSC.
- M. M. El-Metwally, G. M. Abdel-Fattah, F. O. Al-Otibi, D. K. E. Khatieb, Y. A. Helmy, Y. M. Mohammed and W. I. Saber, Heliyon, 2023, 9(9), e20063 CrossRef CAS PubMed.
- W. I. A. Saber, A. A. Ghoniem, F. O. Al-Otibi, M. S. El-Hersh, N. M. Eldadamony, F. Menaa and K. M. Elattar, Sci. Rep., 2023, 13, 13545 CrossRef CAS PubMed.
- J. O. Arawande, A. Akinnusotu and J. O. Alademeyin, Int. J. Tradit. Nat. Med., 2018, 8(1), 13–22 Search PubMed.
- S. Himesh, P. S. Sharan, K. Mishra, N. Govind and A. K. Singhai, Int. Res. J. Pharm., 2011, 2(4), 180–184 Search PubMed.
- A. Amalraj, A. Pius, S. Gopi and S. Gopi, J. Tradit. Complementary Med., 2017, 7(2), 205–233 CrossRef PubMed.
- G. K. Jayaprakasha, L. Jagan Mohan Rao and K. K. Sakariah, J. Agric. Food Chem., 2002, 50(13), 3668–3672 CrossRef CAS PubMed.
- T. Ahmed and A. H. Gilani, Phytother. Res., 2014, 28(4), 517–525 CrossRef CAS PubMed.
- G. D. M. P. Madhusankha, L. F. Siow and Y. Y. Thoo, Food Biosci., 2023, 56, 103171 CrossRef.
- N. M. Umar, T. Parumasivam, N. Aminu and S. M. Toh, J. Appl. Pharm. Sci., 2020, 10(10), 180–194 CAS.
- J. G. de Oliveira Filho, M. J. de Almeida, T. L. Sousa, D. C. dos Santos and M. B. Egea, Bioactive Compounds in Underutilized Vegetables and Legumes, 2021, pp. 297–318 Search PubMed.
- S. Himesh, P. S. Sharan, K. Mishra, N. Govind and A. K. Singhai, Int. Res. J. Pharm., 2011, 2(4), 180–184 Search PubMed.
- L. Labban, Int. J. Pharm. Biomed. Sci., 2014, 5(1), 17–23 Search PubMed.
- A. Bhat, A. M. Mahalakshmi, B. Ray, S. Tuladhar, T. A. Hediyal, E. Manthiannem, J. Padamati, R. Chandra, S. B. Chidambaram and M. K. Sakharkar, BioFactors, 2019, 45(5), 666–689 CrossRef CAS PubMed.
- W. Wongcharoen and A. Phrommintikul, Int. J. Cardiol., 2009, 133(2), 145–151 CrossRef PubMed.
- M. Akaberi, A. Sahebkar and S. A. Emami, Studies on Biomarkers and New Targets in Aging Research in Iran: Focus on Turmeric and Curcumin, 2021, pp. 15–39 Search PubMed.
- H. Van Nong, L. X. Hung, P. N. Thang, V. D. Chinh, L. V. Vu, P. T. Dung, T. Van Trung and P. T. Nga, SpringerPlus, 2016, 5(1), 1–9 CrossRef CAS PubMed.
- (a) K. M. Elattar, A. A. Ghoniem, F. O. Al-Otibi, M. S. El-Hersh, Y. A. Helmy and W. I. Saber, Appl. Sci., 2023, 13(18), 10110 CrossRef CAS; (b) M. M. Hammouda, K. Shalabi, A. A. Alanazi, K. M. Elattar, M. A. Azzam and M. M. Rashed, RSC Adv., 2023, 13(46), 32532–32546 RSC.
- A. Blainski, G. C. Lopes and J. C. P. De Mello, Molecules, 2013, 18(6), 6852–6865 CrossRef CAS PubMed.
- D. Mammen and M. Daniel, Food Chem., 2012, 135(3), 1365–1368 CrossRef CAS PubMed.
- N. J. Miller and C. A. Rice-Evans, Free Radical Res., 1997, 26(3), 195–199 CrossRef CAS PubMed.
- L. L. Mensor, F. N. Menezes, L. C. Leitao, A. S. Reis, T. C. Santos, C. S. Coube and A. G. Oliveira, Phytomedicine, 2001, 8(4), 177–180 Search PubMed.
- I. Mphande, A. Kataba, K. Muzandu and A. Gono-Bwalya, J. Evidence-Based Complementary Altern. Med., 2022, 2022, 7973942, DOI:10.1155/2022/7973942.
- P. Parvekar, J. Palaskar, S. Metgud, R. Maria and S. Dutta, Biomater. Invest. Dent., 2020, 7(1), 105–109 CAS.
- C. Sharma, S. Ansari, M. S. Ansari and S. P. Satsangee, J. Ind. Eng. Chem., 2022, 111, 499–508 CrossRef CAS.
- H. R. El-Seedi, R. M. El-Shabasy, S. A. Khalifa, A. Saeed, A. Shah, R. Shah, F. J. Iftikhar, M. M. Abdel-Daim, A. Omri, N. H. Hajrahand and J. S. Sabir, RSC Adv., 2019, 9(42), 24539–24559 RSC.
- W. S. Cho, F. Thielbeer, R. Duffin, E. M. Johansson, I. L. Megson, W. MacNee, M. Bradley and K. Donaldson, Nanotoxicology, 2014, 8(2), 202–211 CrossRef CAS PubMed.
- M. Zhu, H. Wang, A. A. Keller, T. Wang and F. Li, Sci. Total Environ., 2014, 487, 375–380 CrossRef CAS PubMed.
- D. Luo, C. Yan and T. Wang, Small, 2015, 11(45), 5984–6008 CrossRef CAS PubMed.
- H. Zhang and J. F. Banfield, Chem. Mater., 2005, 17(13), 3421–3425 CrossRef CAS.
- Y. G. Guo, J. S. Lee and J. Maier, Solid State Ionics, 2006, 177(26–32), 2467–2471 CrossRef CAS.
- Y. S. Park, A. Dmytruk, I. Dmitruk, A. Kasuya, Y. Okamoto, N. Kaji, M. Tokeshi and Y. Baba, J. Phys. Chem. C, 2010, 114(44), 18834–18840 CrossRef CAS.
- A. Cartwright, K. Jackson, C. Morgan, A. Anderson and D. W. Britt, Agronomy, 2020, 10(7), 1018 CrossRef CAS.
- A. Al-Asfar, Z. Zaheer and E. S. Aazam, J. Photochem. Photobiol., B, 2018, 185, 143–152 CrossRef CAS PubMed.
- S. W. Im, H. Y. Ahn, R. M. Kim, N. H. Cho, H. Kim, Y. C. Lim, H. E. Lee and K. T. Nam, Adv. Mater., 2020, 32(41), 1905758 CrossRef CAS PubMed.
- Q. Zhang, X. Yang, H. Ji, D. Sun and Y. Liu, ACS Appl. Mater. Interfaces, 2013, 5(19), 9355–9362 CrossRef PubMed.
- (a) H. Xu, F. Wang and Y. Zhu, J. Colloid Interface Sci., 2012, 377, 244–250 CrossRef PubMed; (b) M. M. Hammouda, M. M. Rashed, K. M. Elattar and A. M. Osman, RSC Adv., 2023, 13(17), 11600–11634 RSC.
- M. S. El-Hersh, W. Saber, H. A. El-Fadaly and M. K. Mahmoud, Pak. J. Biol. Sci., 2016, 19, 279–288 CrossRef CAS PubMed.
- H.-L. Lau, F. W. F. Wong, R. N. Z. R. Abd Rahman, M. S. Mohamed, A. B. Ariff and S.-L. Hii, Biocatal. Agric. Biotechnol., 2023, 50, 102696 CrossRef CAS.
- A. Elsayed, Z. Moussa, S. S. Alrdahe, M. M. Alharbi, A. A. Ghoniem, A. Y. El-Khateeb and W. I. Saber, Front. Microbiol., 2022, 13, 893603 CrossRef PubMed.
- R. Pandiselvam, V. Prithviraj, M. Manikantan, P. S. Beegum, S. Ramesh, S. Padmanabhan, A. Kothakota, A. Mathew, K. Hebbar and A. M. Khaneghah, Measurement: Food, 2022, 5, 100015 Search PubMed.
- V. Srikanth, G. Rajesh, A. Kothakota, R. Pandiselvam, N. Sagarika, M. Manikantan and K. Sudheer, Computers and Electronics in Agriculture, 2020, 177, 105715 CrossRef.
- K. T. Beier, E. E. Steinberg, K. E. DeLoach, S. Xie, K. Miyamichi, L. Schwarz, X. J. Gao, E. J. Kremer, R. C. Malenka and L. Luo, Cell, 2015, 162(3), 622–634 CrossRef CAS PubMed.
- Z. Moussa, D. B. Darwish, S. S. Alrdahe and W. I. Saber, Front. Microbiol., 2021, 12, 731262 CrossRef PubMed.
- W. I. Saber, N. E.-A. El-Naggar, M. S. El-Hersh, A. Y. El-Khateeb, A. Elsayed, N. M. Eldadamony and A. A. Ghoniem, Sci. Rep., 2021, 11, 1–15 CrossRef PubMed.
- A. A. Al-Askar, E. M. Rashad, Z. Moussa, K. M. Ghoneem, A. A. Mostafa, F. O. Al-Otibi, A. A. Arishi and W. I. Saber, Front. Microbiol., 2022, 13, 772417 CrossRef PubMed.
- M. G. Anjaly, M. Prince, A. S. Warrier, A. N. Lal, N. K. Mahanti, R. Pandiselvam, R. Thirumdas, R. Sreeja, A. V. Rusu and M. Trif, Ultrason. Sonochem., 2022, 90, 106166, DOI:10.1016/j.ultsonch.2022.106166.
- M. Sarkar, S. Denrah, M. Das and M. Das, Chemical Physics Impact, 2021, 3, 100053 CrossRef.
- N. Liaqat, N. Jahan, T. Anwar and H. Qureshi, Front. Chem., 2022, 10, 952006 CrossRef CAS PubMed.
- D. Singh, V. Rathod, S. Ninganagouda, J. Hiremath, A. K. Singh and J. Mathew, Bioinorg. Chem. Appl., 2014, 2014, 408021, DOI:10.1155/2014/408021.
- B. A. El-deeb, E. Asem and K. Mohammed, Sohag Journal of Sciences, 2023, 8(1), 1–6 CrossRef.
- S. S. Birla, S. C. Gaikwad, A. K. Gade and M. K. Rai, Sci. World J., 2013, 2013, 796018, DOI:10.1155/2013/796018.
- W. I. Saber, N. E. El-Naggar, M. S. El-Hersh and A. Y. El-Khateeb, Biotechnology, 2015, 14, 47–57 CrossRef CAS.
- M. S. Elsayed, N. M. Eldadamony, S. S. Alrdahe and W. I. Saber, Molecules, 2021, 26, 5048 CrossRef CAS PubMed.
- L. E Wright, J. B. Frye, B. Gorti, B. N. Timmermann and J. L. Funk, Curr. Pharm. Des., 2013, 19(34), 6218–6225 CrossRef PubMed.
- J. N. Jacob, Stud. Nat. Prod. Chem., 2016, 48, 101–135 CAS.
- J. O. Arawande, A. Akinnusotu and J. O. Alademeyin, Int. J. Tradit. Nat. Med., 2018, 8(1), 13–22 Search PubMed.
- L. J. Jing, M. Mohamed, A. Rahmat and M. F. A. Bakar, J. Med. Plants Res., 2010, 4(1), 27–32 CAS.
- K. I. Priyadarsini, Molecules, 2014, 19(12), 20091–20112 CrossRef PubMed.
- A. Arcadi, Chem. Rev., 2008, 108(8), 3266–3325 CrossRef CAS PubMed.
- K. Hariharan, P. Patel and T. Mehta, Pharm. Dev. Technol., 2022, 27(6), 665–683 CrossRef CAS PubMed.
- S. Phukan, P. Bharali, A. K. Das and M. H. Rashid, RSC Adv., 2016, 6(55), 49307–49316 RSC.
- H. S. Lim, S. H. Park, K. Ghafoor, S. Y. Hwang and J. Park, Food Chem., 2011, 124(4), 1577–1582 CrossRef CAS.
- A. Amalraj, A. Pius, S. Gopi and S. Gopi, J. Tradit. Complementary Med., 2017, 7(2), 205–233 CrossRef PubMed.
- M. Yousefian, N. Shakour, H. Hosseinzadeh, A. W. Hayes, F. Hadizadeh and G. Karimi, Phytomedicine, 2019, 55, 200–213 CrossRef CAS PubMed.
- Y. Xue, Q. Luan, D. Yang, X. Yao and K. Zhou, J. Phys. Chem. C, 2011, 115(11), 4433–4438 CrossRef CAS.
- S. Patil, A. Sandberg, E. Heckert, W. Self and S. Seal, Biomaterials, 2007, 28(31), 4600–4607 CrossRef CAS PubMed.
- Q. Mu, G. Jiang, L. Chen, H. Zhou, D. Fourches, A. Tropsha and B. Yan, Chem. Rev., 2014, 114(15), 7740–7781 CrossRef CAS PubMed.
- M. Horie and Y. Tabei, Free Radical Res., 2021, 55(4), 331–342 CrossRef CAS PubMed.
- D. Zheng, C. Huang, H. Huang, Y. Zhao, M. R. U. Khan, H. Zhao and L. Huang, Chem. Biodiversity, 2020, 17(8), e2000171 CrossRef CAS PubMed.
- F. Z. Gul and M. Basheer, J. Ayurvedic Herb. Med., 2016, 2(5), 192–199 CrossRef.
- T. Ak and I. Gülçin, Chem.-Biol. Interact., 2008, 174(1), 27–37 CrossRef CAS PubMed.
- F. Nazzaro, F. Fratianni, A. d'Acierno, V. De Feo, F. J. Ayala-Zavala, A. Gomes-Cruz, D. Granato and R. Coppola, Effect of polyphenols on microbial cell-cell communications, in Quorum Sensing, Academic Press, 2019, pp. 195–223 Search PubMed.
- B. Khalandi, N. Asadi, M. Milani, S. Davaran, A. J. N. Abadi, E. Abasi and A. Akbarzadeh, Drug Res., 2017, 11(02), 70–76 Search PubMed.
- L. Z. Flores-López, H. Espinoza-Gómez and R. Somanathan, J. Appl. Toxicol., 2019, 39(1), 16–26 CrossRef PubMed.
- B. Khalandi, N. Asadi, M. Milani, S. Davaran, A. J. N. Abadi, E. Abasi and A. Akbarzadeh, Drug Res., 2017, 11(02), 70–76 Search PubMed.
- J. S. Kim, E. Kuk, K. N. Yu, J. H. Kim, S. J. Park, H. J. Lee, S. H. Kim, Y. K. Park, Y. H. Park, C. Y. Hwang and Y. K. Kim, Nanomedicine, 2007, 3(1), 95–101 CrossRef CAS PubMed.
- S. Ninganagouda, V. Rathod, H. Jyoti, D. Singh, K. Prema and M. U. Haq, Int. J. Pharma Bio Sci., 2013, 4(2), 222–229 CAS.
- P. S. Murthy, A. Das, B. Ramalingam, R. Sekar and S. Dobretsov, Nanotechnology Approaches to Treating Antimicrobial Resistant Infections Caused by Biofilms: Research Needs for Translation from In Vitro to In Vivo Applications, in Biofilm Associated Antimicrobial Resistance and Its Recovery, CRC Press, 2013, pp. 116–158 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra00004h |
| This journal is © The Royal Society of Chemistry 2024 |