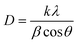Open Access Article
Open Access ArticleFacile sonochemically-assisted bioengineering of titanium dioxide nanoparticles and deciphering their potential in treating breast and lung cancers: biological, molecular, and computational-based investigations†
Nada K. Sedky‡
 a,
Noha Khalil Mahdy‡b,
Nour M. Abdel-kaderac,
Manal M. M. Abdelhadyd,
Mohamad Maged
a,
Noha Khalil Mahdy‡b,
Nour M. Abdel-kaderac,
Manal M. M. Abdelhadyd,
Mohamad Maged e,
Aya L. Allamf,
Mohammad Y. Alfaifig,
Samir N. Shammah,
Hatem A. F. M. Hassan
e,
Aya L. Allamf,
Mohammad Y. Alfaifig,
Samir N. Shammah,
Hatem A. F. M. Hassan fi and
Sherif Ashraf Fahmy
fi and
Sherif Ashraf Fahmy *j
*j
aDepartment of Biochemistry, School of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, R5 New Garden City, New Administrative Capital, Cairo 11835, Egypt
bDepartment of Pharmaceutics and Industrial Pharmacy, Faculty of Pharmacy, Cairo University, Kasr El-Aini Street, Cairo 11562, Egypt
cDepartment of Biochemistry, Faculty of Science, Ain Shams University, Cairo 11566, Egypt
dClinical Pharmacy Department, Faculty of Pharmacy, Badr University, Cairo 11829, Egypt
eFaculty of Biotechnology, Nile University, Giza, Egypt
fSchool of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, New Administrative Capital, Egypt
gKing Khalid University, Faculty of Science, Biology Department, Abha 9004, Saudi Arabia
hInstitute of Global Health and Human Ecology, School of Sciences & Engineering, The American University in Cairo, AUC Avenue, P.O. Box 74, New Cairo 11835, Egypt
iDepartment of Pharmaceutics and Industrial Pharmacy, Faculty of Pharmacy, Cairo University, 11562 Cairo, Egypt
jDepartment of Chemistry, School of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, R5 New Garden City, New Capital, Cairo 11835, Egypt. E-mail: sheriffahmy@aucegypt.edu; Tel: +20-1222613344
First published on 14th March 2024
Abstract
Combining sonochemistry with phytochemistry is a modern trend in the biosynthesis of metallic nanoparticles (NPs), which contributes to the sustainability of chemical processes and minimizes hazardous effects. Herein, titanium dioxide (TiO2) NPs were bioengineered using a novel and facile ultrasound-assisted approach utilizing the greenly extracted essential oil of Ocimum basilicum. FTIR and UV-Vis spectrophotometry were used to confirm the formation of TiO2 NPs. The X-ray diffraction (XRD) analysis showed the crystalline nature of TiO2 NPs. TEM analysis revealed the spherical morphology of the NPs with sizes ranging from 5.55 to 13.89 nm. Energy-dispersive X-ray (EDX) confirmed the purity of the greenly synthesized NPs. TiO2 NPs demonstrated outstanding antitumor activity against breast (MCF-7) and lung (A-549) cancer cells with estimated IC50 values of 1.73 and 4.79 μg mL−1. The TiO2 NPs were cytocompatible to normal cells (MCF-10A) with a selectivity index (SI) of 8.77 for breast and 3.17 for lung cancer. Biological assays revealed a promising potential for TiO2 NPs to induce apoptosis and arrest cells at the sub-G1 phase of the cell cycle phase in both cancer cell lines. Molecular investigations showed the ability of TiO2 NPs to increase apoptotic genes' expression (Bak and Bax) and their profound ability to elevate the expression of apoptotic proteins (caspases 3 and 7). Molecular docking demonstrated strong binding interactions for TiO2 NPs with caspase 3 and EGFR-TK targets. In conclusion, the greenly synthesized TiO2 NPs exhibited potent antitumor activity and mitochondrion-based cell death against breast and lung cancer cell lines while maintaining cytocompatibility against normal cells.
1. Introduction
Cancer is one of the leading causes of mortality in the world. It poses a burden on societies, healthcare systems, and the quality of human life.1–3 One in every six deaths is attributed to cancer globally. Breast and lung tumors are at the top of the list as the world's most prevalent cancers.4,5 Global efforts are needed to find novel chemotherapeutic agents or enhance the efficacy and safety of existing drugs.5 Some tumors fail to respond to treatment owing to the poor absorption of the chemotherapeutic drug into the targeted cells6 or extensive adverse effects on normal cells.7,8 The advent of nanotechnology paved the way for cancer research to explore more antitumor treatment agents; thanks to the “enhanced permeability and retention” (EPR) phenomenon that enabled the passive targeting of nanosized particles into the tumor cells via their leaky distorted vasculature system that is not typically observed in healthy cells.7,9–11 Nanoparticle delivery systems transport the antitumor drug preferentially into cancer cells due to their extensive blood supply and leaky cell membranes.8,12 Interestingly, antitumor drugs are retained inside cancer cells because of the restricted lymphatic drainage.13,14 Metallic nanoparticles, such as Ag, Au, Cu, Zn, Ti and Pt NPs antineoplastic agents due to their inert behavior towards normal cells, yet selective-targeting to the leaky cancer membranes.7,15–17 Thus, their application as potentially effective yet safe anticancer agents has drawn much attention recently.18In particular, titanium dioxide nanoparticles (TiO2 NPs) have shown valuable applicability in various fields, including cosmetics, sericulture, wastewater remediation, air purification, and the food industry.19 Smart improvements in their synthesis and formulation were necessary to overcome some of their chemical instability in aqueous solutions.20 TiO2 NPs have distinguished properties relevant to other metals. Besides, TiO2 NPs have promising biomedical applications in treating bacterial and viral infections. In addition, TiO2 NPs were tested as nanocarriers for several antitumor drugs.21,22 But not until recently they were shown to serve as potential antitumor agents by themselves against solid cancers.23 TiO2 NPs rose as promising candidates in the antitumor arsenal due to their remarkable surface properties, chemical stability, biocompatibility, and biological anti-proliferative activity,15 as compared to other metallic NPs. Several chemical methods have been utilized to synthesize different metallic NPs. However, these methods involve toxic solvents with several perilous environmental and human health implications.24 Thus, numerous green approaches have been developed for synthesizing various metallic NPs and were considered a breakthrough in enhancing the activity of metallic NPs while minimizing toxicity and environmentally hazardous effects.14 Green synthesis entails using plant extracts or microorganisms rather than harsh chemicals. These eco-friendly methods are biocompatible, preventive to unnecessary wastes, cost-effective, and safe for the environment and humans.25 It relies on various phytochemicals, such as polysaccharides, terpenoids, organic acids, proteins etc.,26 to reduce metallic atoms, converting them into nanoscale counterparts (bottom-up approach).25,27,28 In addition, natural compounds aid in stabilizing the metallic NPs and enhancing their surface properties.24,29
Ocimum basilicum, basil, is a well-known herb that possesses numerous potential medicinal and biotechnological applications. Natural ingredients extracted from basil were reported to have antibacterial and anticancer activities.27,30–33 Some efforts were committed to synthesizing metal nanoparticles using basil herbal extract. However, to the best of our knowledge, no studies in the literature reported the biofabrication of anticancer TiO2 NPs using basil essential oils.30,34 Most of the previous studies focused on the antibacterial activity35,36 rather than the antitumor effect of basil-mediated-TiO2 NPs.
In this study, we designed a novel eco-friendly TiO2 NPs using a greenly extracted essential oil of O. basilicum. The greenly engineered NPs were characterized physicochemically using various techniques, including spectroscopic techniques, X-ray diffraction, transmission electron microscopy, scanning Electron Microscopy (SEM), and Energy-Dispersive X-ray (EDX). The greenly synthesized nanoparticles were then evaluated for their biological activity using a comprehensive panel of cellular biology, bioinformatics, cytogenetics, molecular biology, and molecular docking techniques. Several assays were performed, including apoptosis, cell cycle analysis, gene expression analysis, and western blotting, to examine the underlying molecular mechanisms of our green TiO2 NPs-induced cell death in both tumor models. In addition, a molecular docking study was conducted to predict the binding interactions of TiO2 NPs with two of the most well-known targets in both breast and lung tumors: caspase 3 and EGFR-TK.
2. Methods
2.1. Green extraction and characterization of Ocimum basilicum essential oil (OBO)
OBO was extracted using a green method by hydrodistillating the aerial parts of O. basilicum L. locally grown in Greece, at the whole flowering stage, utilizing a Clevenger-type apparatus. Hydrodistillation was carried out by adding 250 g of fresh plant material to 1 L of distilled water. The hydrodistillation took place at 110 °C, which was then reduced to 80 °C, keeping the cooling system's temperature at 4 °C. The obtained OBO was then stored in firmly closed amber glass bottles at 4 °C for further experiments. The chemical components of the extracted oil were analyzed using GC-MS (Agilent Technologies gas chromatography (7890B) and a mass spectrometer detector (5977B)), as detailed in our previous studies.14,372.2. Sonochemical green synthesis of TiO2 NPs
Eco-friendly TiO2 NPs were synthesized by reducing titanium(I) isopropoxide using OBO and sonochemical reaction. In brief, 5 mL of OBO was added to 15 mL of titanium(I) isopropoxide solution (10 mM) while being ultrasound irradiated using an ultrasonic homogenizer (Pulse 150 Benchmark, USA) for 15 min. A horn of 21 kHz frequency was placed centrally at 1 cm depth of the mixture with an optimum sonication amplitude of 40%. The obtained TiO2 NPs were purified by ultracentrifugation for 30 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, and the remaining pellet was washed thrice with deionized water to remove any plant residual materials. The final pellet was resuspended in deionized water and then subjected to freeze drying for 24 h.
000 rpm, and the remaining pellet was washed thrice with deionized water to remove any plant residual materials. The final pellet was resuspended in deionized water and then subjected to freeze drying for 24 h.
2.3. Characterization of the eco-friendly TiO2 NPs
In this equation, D is the crystallite diameter (nm), k is the Scherrer constant and is equal to 0.9, λ is the X-ray Cu Kα radiation wavelength (°), θ is the peak position (radians), and β is the full width at half-maximum (radians).38
2.4. Cell culture
Human A-549 (lung cancer) and MCF-7 (Breast Adenocarcinoma) were used as cancer models, while the control used was MCF-10A (normal breast epithelium). Cell lines were provided by the American type of culture collection (ATCC, Wesel, Germany). Corresponding cells were cultured in standard DMEM media containing 100 units per mL penicillin and 100 mg mL−1 streptomycin. The media was supplemented with10% heat-inactivated fetal bovine serum. Incubation was done at a humidified 5% (v/v) CO2 atmosphere, and the temperature was set to 37 °C.2.5. Cell viability assay
Cell viability assay was done using the specified SRB approach.39 100 μL cell suspension aliquots containing (5 × 103 cells) were placed in 96-well plates and grown in the designated media for 24 hours. Treated cells were exposed to 100 μL media containing TiO2 NPs at different concentrations (0.03, 0.3, 3, 30, 300 μg mL−1). Following 48 hours of drug exposure, cell fixation was routinely done by substituting the media with 150 μL 10% TCA and incubating for 1 hour at 4 °C. Then TCA solution was aspirated, and cells were washed 5 times with distilled water. A 70 μL aliquots of SRB solution (0.4% w/v) were put and incubated in the dark for 10 min at ambient room temperature. Wells were washed with 1% acetic acid 3 times and left to air-dry overnight. To dissolve protein-bound SRB stain, 150 μL of 10 mM TRIS was accordingly added; BMGLABTECH®-FLUOstar Omega microplate reader (Ortenberg, Germany) set at 540 nm was used to measure absorbance.2.6. Apoptosis
Annexin V-FITC apoptosis detection kit (Abcam Inc., Cambridge Science Park, Cambridge, UK), was used to calculate apoptosis or necrosis cell populations according to the recommended protocol. Measurements were recorded using 2 fluorescent-channels flow cytometry. TiO2 NPs treatment was completed at the defined IC50 concentrations for 48 hours, followed by cells (105 cells) collection. Routine trypsinization was done to loosen up cells, followed by washing with ice-cold PBS (pH 7.4) 2 times. Then, 0.5 mL of Annexin V-FITC/PI solution was added to the cells and incubated in the dark for 30 minutes according to the manufacturer's protocol. Cells were stained and then injected via ACEA Novocyte™ flowcytometer (ACEA Biosciences Inc., San Diego, CA, USA). FITC and PI fluorescent signals were analyzed (λex/em 488/530 nm for FITC and λex/em 535/617 nm for PI, respectively) using FL1 and FL2 signal detector. An estimate of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events, for each sample, were captured. Quantification of positive FITC and/or PI cells was achieved by quadrant analysis and estimated by ACEA NovoExpress™ software (ACEA Biosciences Inc., San Diego, CA, USA).
000 events, for each sample, were captured. Quantification of positive FITC and/or PI cells was achieved by quadrant analysis and estimated by ACEA NovoExpress™ software (ACEA Biosciences Inc., San Diego, CA, USA).
2.7. Cell cycle analysis
MCF-7 and A-549 cell lines were treated with TiO2 NPs at the determined IC50s for 48 hours, 105 cells were harvested for routine trypsinization. Washing was done twice with ice-cold PBS (pH 7.4). Resuspension of cells was achieved by adding 2 mL 60% ice-cold ethanol. Cell's fixation was done by incubation for 1 hour at 4 °C. Then, washing of fixed cells was done using PBS (pH 7.4) twice and eventually resuspended in 1 mL solution (PBS pH 7.4 containing 10 μg mL−1 propidium iodide (PI) and 50 μg mL−1 RNAase A). Incubation was done at 37 °C for 20 minutes. Flow cytometry was used to analyze DNA content by FL2 (λex/em 535/617 nm) signal detector (ACEA Novocyte™ flow cytometer, ACEA Biosciences Inc., San Diego, CA, USA). An estimate of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events, per sample were acquired. ACEA NovoExpress™ software (ACEA Biosciences Inc., San Diego, CA, USA) was eventually used to evaluate cell cycle distribution.
000 events, per sample were acquired. ACEA NovoExpress™ software (ACEA Biosciences Inc., San Diego, CA, USA) was eventually used to evaluate cell cycle distribution.
2.8. Gene selection based on bioinformatics' analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC| > 0.1 and adj-p < 0.05 in the DESeq2 R package.
FC| > 0.1 and adj-p < 0.05 in the DESeq2 R package.2.9. Gene expression for apoptotic markers
Total cellular RNA was extracted using the QIA amp Viral RNA Mini Kit (Qiagen, Hilden, Germany) as per the manufacturer's guidelines. ReverAid RT Kit purchased from (ThermoFisher Scientific, Waltham, USA) and Bio-Rad™ 100 thermal cycler were used to reverse transcribe the isolated RNA according to the kit's protocol. 1 μL of cDNA collected from each sample was assayed afterwards by quantitative real-time PCR (RT-qPCR) for the target genes, BAX, BAK and the housekeeping β-actin gene. The primers' sequences used in this study for the RT-qPCR analysis are reported in Table S1.† The assay was done on a “Rotor-Gene Q-Qiagen Real-time PCR thermal cycler” using the standard conditions. The resulting data were normalized to the control β-actin, and the relative normalized gene expression output was measured by the 2−ΔΔCt approach.402.10. Immunoblotting for apoptotic proteins
Western blot analysis was deployed to determine the relative quantity of caspase-3 and caspase-7 proteins in the cells treated with TiO2 NPs as compared to untreated cells (control). The β-actin protein was chosen for data normalization. Briefly, MCF-7 cells were scraped off the plate using a cold, sterile cell scraper and transferred into a pre-cooled microcentrifuge tube where agitation was performed for 30 min at 4 °C. Whenever necessary, cells were sonicated 3 times for 10–15 s to complete cell lysis. The cellular solution was then spun at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min in a 4 °C pre-cooled centrifuge. The supernatant was translocated to a fresh tube while the pellet was discarded. A small volume (20 μL) of our lysate was allocated to the protein assay. The protein concentration was measured for each cell lysate. The 2× Laemmli sample buffer was then added at an equal volume to that of the sample and heated to 95 °C for 5 min. This was followed by a centrifugation step at 16
000g for 20 min in a 4 °C pre-cooled centrifuge. The supernatant was translocated to a fresh tube while the pellet was discarded. A small volume (20 μL) of our lysate was allocated to the protein assay. The protein concentration was measured for each cell lysate. The 2× Laemmli sample buffer was then added at an equal volume to that of the sample and heated to 95 °C for 5 min. This was followed by a centrifugation step at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g in a microcentrifuge for 1 min. At this moment, the samples were prepared and ready to be loaded into the SDS (mini SDS-PAGE gel), where equal amounts of protein (25 μg) were loaded into the wells, along with molecular weight markers (ThermoScientific PageRuler Broad Range Unstained Protein Ladder, Cat #: 26630). The gel was left to run for 5 min at 90 V, then the voltage was increased to 100–150 V to speed up the run. Directly afterwards, the gel was placed in 1× transfer buffer for 10–15 min. The transfer sandwich was then prepared, ensuring no air bubbles entered the assembly. The blot was allocated to the cathode, and the gel was placed at the anode. The cassette was moved to the transfer tank containing ice blocks. Accordingly, the protein was transferred from the gel to the membrane. The sample was blocked in 3% bovine serum albumin (BSA) in Tris-buffered saline with Tween 20 (TBST) buffer at room temperature for 1 h. The samples were incubated with the primary antibodies against the target proteins (Table S2†) overnight at 4 °C. The blot was then washed 3–5 times for 5 min with TBST and incubated with HRP-conjugated secondary antibodies (Table S2†) for 1 h at room temperature. The blot was then rinsed 3–5 times for 5 min with TBST. According to the manufacturer's guidelines, the chemiluminescent reaction was done using the ECL western blot HRP substrate (Pierce, ThermoFisher Scientific). The chemiluminescent signals were captured using the ChemiDoc imaging system of Bio-Rad. This software was also used in the densitometry calculations.
000g in a microcentrifuge for 1 min. At this moment, the samples were prepared and ready to be loaded into the SDS (mini SDS-PAGE gel), where equal amounts of protein (25 μg) were loaded into the wells, along with molecular weight markers (ThermoScientific PageRuler Broad Range Unstained Protein Ladder, Cat #: 26630). The gel was left to run for 5 min at 90 V, then the voltage was increased to 100–150 V to speed up the run. Directly afterwards, the gel was placed in 1× transfer buffer for 10–15 min. The transfer sandwich was then prepared, ensuring no air bubbles entered the assembly. The blot was allocated to the cathode, and the gel was placed at the anode. The cassette was moved to the transfer tank containing ice blocks. Accordingly, the protein was transferred from the gel to the membrane. The sample was blocked in 3% bovine serum albumin (BSA) in Tris-buffered saline with Tween 20 (TBST) buffer at room temperature for 1 h. The samples were incubated with the primary antibodies against the target proteins (Table S2†) overnight at 4 °C. The blot was then washed 3–5 times for 5 min with TBST and incubated with HRP-conjugated secondary antibodies (Table S2†) for 1 h at room temperature. The blot was then rinsed 3–5 times for 5 min with TBST. According to the manufacturer's guidelines, the chemiluminescent reaction was done using the ECL western blot HRP substrate (Pierce, ThermoFisher Scientific). The chemiluminescent signals were captured using the ChemiDoc imaging system of Bio-Rad. This software was also used in the densitometry calculations.
2.11. Molecular docking
Titanium dioxide NPs (TiO2 NPs) activity against cell proliferation and molecular pro-apoptotic activity was tested in silico by predicting its ability to activate the caspase cascade of A-549 and MCF-7 cancer cell lines upon binding with caspase-3. The X-ray crystallography derived caspase-3 (CAS329306) in complex with 4-methyl-benzenesulfonamide (MB) was obtained from Protein Data Bank (PDB ID: 2XYG; http://www.rcsb.org/structure/2XYG).41 GFR-tyrosine kinases (EGFR-TK) were also chosen as a vital receptor in triggering and generating signaling reactions for both cell lung cancer (CLC) and breast cancer (BC) (PDB ID: 1M17; http://www.rcsb.org/structure/1M17).42,43Water molecules in the 2 crystal structures were deleted using AutoDock tools, and polar hydrogen atoms were added. Marsili–Gasteiger partial charges are appointed employing a two-phase algorithm. The receptor crystal structures were subjected to energy minimization using the AutoDockTools (ADT, v1.5.6) prepare_receptor4.py command, where Kollman-united charge was used for calculating the partial atomic charge.
The structure of TiO2 was drawn using ChemSketch, and its geometry was optimized by Autodock 4.2. To determine the preferred binding sites on crystal structure, the grid box center was set to (x = 36.357, y = 38.829, z = 32.088) and (x = 20.091, y = −1.872, z = 58.436) for PDB IDs 2XYG and 1M17, respectively. The output from AutoDock was further analyzed with UCSF Chimera and PyMOL software package.44
2.12. Statistical analysis
Data analysis and visualization were accomplished with the aid of GraphPad Prism 6. All the included assays were performed in triplicates. Accordingly, the results are presented as the mean of three individual runs ± standard deviation (SD). The Shapiro–Wilk Test was performed to ensure the normal distribution of the data obtained. Regarding the comparisons of the two groups, a Student's t-test was used. As per the comparisons that include three or more groups, ANOVA with multiple comparisons post hoc test was used. Statistical significance is considered at P-value ≤ 0.05.3. Results and discussion
Breast and lung cancers are two leading causes of death globally.4 Together, they account for 30% of total cancer deaths.1 In addition, the lung is a very likely site for breast cancer metastasis.45 Thus, there is a pressing need to develop eco-friendly antitumor agents that can target breast and lung cancers using facile green approaches. In this regard, we greenly synthesized OBO-mediated TiO2 nanoparticles using a sonochemical approach. OBO is prosperous with several bioreductants and capping agents, as evident from the GC-LC analysis, thus, it was exploited for the one-step facile green synthesis of TiO2 NPs with the help of ultrasonic waves.31,46–483.1. Chemical analysis of the extracted OBO using gas chromatography-mass spectrometry (GC-MS)
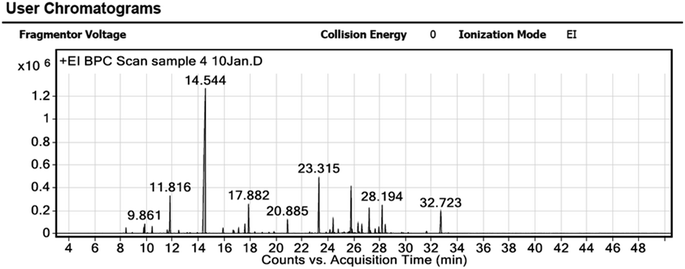
The GC-MS chromatogram of the greenly extracted OBO is presented in Fig. 1, showing the presence of 58 compounds. The mass spectra of the constituents are listed in Table 1 after being identified with the National Institute of Standards and Technology (NIST) library. Linalool, a monoterpenoid, was recognized as the major component (46.55%) of the analyzed oil. Eugenol, a phenylpropanoid (8.27%), cis-α-bergamotene, a bicyclic sesquiterpenoid (6.25%); and Eucalyptol, a monoterpenoid (4.53%) were the second, third and fourth most prevalent components, respectively. These compounds were reported to have potent antioxidant activity, and thus, they were the key players in the bioreduction of titanium(I) isopropoxide into TiO2 NPs.49,50 Moreover, the GC-MS analysis of the extracted Ocimum basilicum oil established the presence of other terpene compounds that serve as natural reducing agents. These results are in line with previously reported studies.51,52| Peak | RT | Compound | Area sum (%) |
|---|---|---|---|
| 1 | 8.413 | α-Pinene | 0.51 |
| 2 | 8.897 | Camphene | 0.07 |
| 3 | 9.782 | Sabinene | 0.51 |
| 4 | 9.861 | 2-β-Pinene | 1 |
| 5 | 10.07 | 1-Octen-3-ol | 0.06 |
| 6 | 10.433 | β-Myrcene | 0.72 |
| 7 | 11.597 | p-Cymene | 0.37 |
| 8 | 11.816 | Eucalyptol | 4.53 |
| 9 | 12.486 | β-Ocimene | 0.33 |
| 10 | 13.138 | cis-Sabinene hydrate | 0.09 |
| 11 | 13.348 | Linalool oxide B | 0.11 |
| 12 | 13.93 | α-Terpineol | 0.12 |
| 13 | 14.544 | Linalool | 46.55 |
| 14 | 14.609 | Hotrienol | 0.05 |
| 15 | 15.908 | 2-Bornanone | 0.72 |
| 16 | 16.695 | endo-Borneol | 0.38 |
| 17 | 16.751 | α-Terpineol | 0.25 |
| 18 | 17.114 | Terpinen-4-OL | 0.6 |
| 19 | 17.598 | L-α-Terpineol | 1.11 |
| 20 | 17.882 | Estragole | 3.44 |
| 21 | 18.361 | n-Octyl acetate | 0.16 |
| 22 | 18.934 | β-Citronellol | 0.1 |
| 23 | 19.446 | Carvone | 0.13 |
| 24 | 19.842 | cis-Geraniol | 0.22 |
| 25 | 20.885 | Iso-bornyl acetate | 1.66 |
| 26 | 22.602 | Elixene | 0.13 |
| 28 | 23.315 | Eugenol | 8.27 |
| 29 | 23.873 | α-Cubebene | 0.15 |
| 30 | 24.162 | (−)-β-Bourbonene | 0.49 |
| 31 | 24.409 | β-Elemene | 1.8 |
| 32 | 24.52 | α-Copaene | 0.08 |
| 33 | 24.8 | Methyleugenol | 0.42 |
| 34 | 25.121 | trans-Sesquisabinene hydrate | 0.06 |
| 35 | 25.261 | Isocaryophillene | 0.18 |
| 36 | 25.568 | γ-Muurolene | 0.12 |
| 37 | 25.652 | β-Gurjunene | 0.2 |
| 38 | 25.796 | cis-α-Bergamotene | 6.2 |
| 39 | 25.875 | α-Guaiene | 0.44 |
| 40 | 25.992 | (+)-β-Funebrene | 0.06 |
| 42 | 26.331 | Humulene | 1.25 |
| 43 | 26.42 | cis-β-Farnesene | 0.17 |
| 44 | 26.625 | (+)-epi-Bicyclosesquiphellandrene | 1.09 |
| 45 | 27.193 | Germacrene-D | 3.24 |
| 46 | 27.281 | cis-β-Farnesene | 0.35 |
| 47 | 27.654 | γ-Elemene | 0.64 |
| 48 | 27.938 | δ-Guaiene | 0.83 |
| 49 | 28.194 | γ-Cadinene | 3.55 |
| 50 | 28.436 | cis-Calamenene | 1.13 |
| 52 | 28.878 | α-Cadinene | 0.07 |
| 53 | 29.707 | (±)-trans-Nerolidol | 0.11 |
| 55 | 30.209 | (+)-Spathulenol | 0.11 |
| 56 | 31.615 | Cubenol | 0.34 |
| 57 | 32.723 | α-Cadinol | 4.41 |
| 58 | 33.305 | α-epi-Muurolol | 0.06 |
3.2. Characterization of the eco-friendly TiO2 NPs
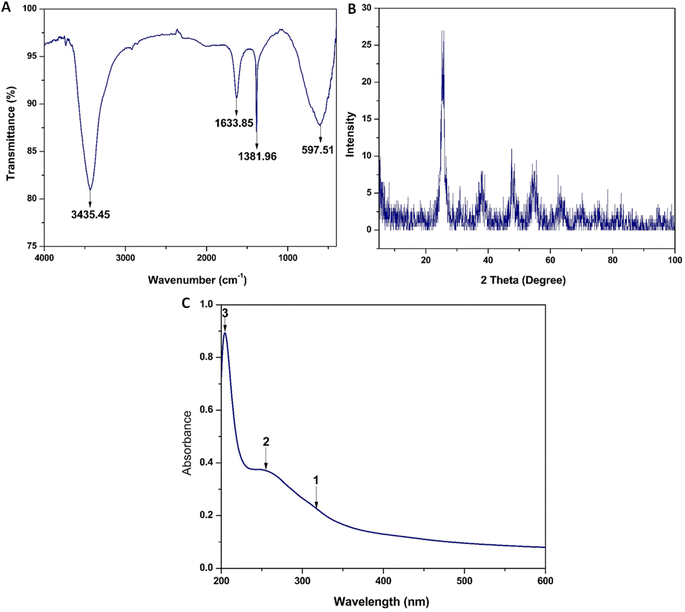
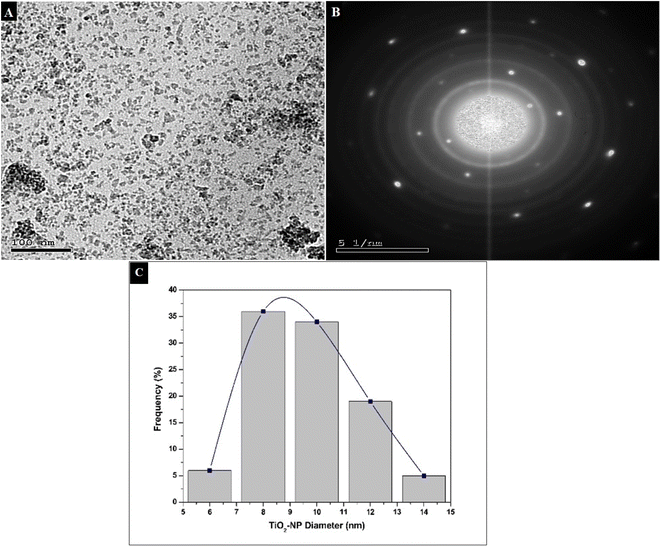
TiO2 NPs were synthesized using a green and facile method that was environmentally friendly, cheap, and did not involve hazardous reagents. Unlike the chemical methods, our simple method is more effective and benign because of the use of green sonochemical reduction combined with the natural reductants from the greenly extracted OBO to generate the TiO2 NPs. Sonochemical green synthesis is an ultrasound-assisted approach that relies on acoustic cavitation without directly impacting the bond's vibrational energy, resulting in nano-sized particles with high surface area and optimized morphologies and reactivity.53The prepared TiO2 NPs were characterized with regard to their physicochemical properties to confirm the successful synthesis of the crystalline NPs.
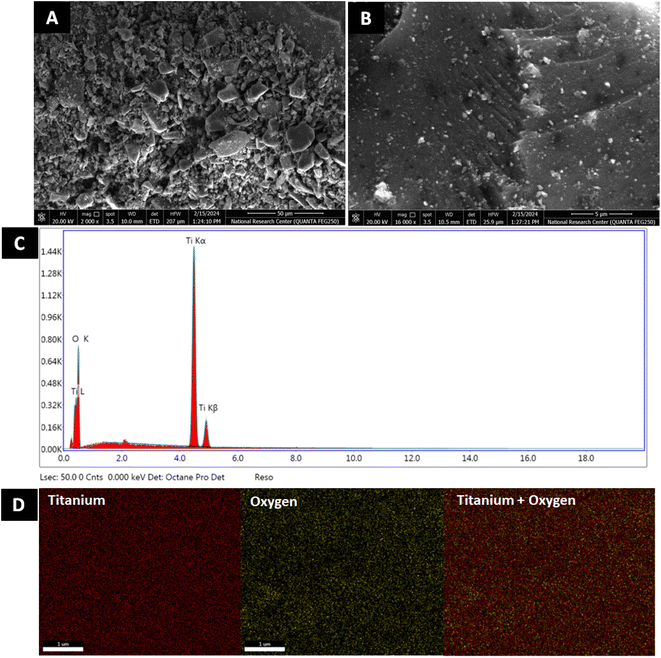 | ||
| Fig. 3 Morphological and structural analysis of TiO2 NPs. SEM of TiO2 NPs at (A) 50 μm and (B) 5 μm, (C) EDX profile and (D) EDX mapping analysis of TiO2 NPs. | ||
| Sample | Element | Weight% | Atomic% |
|---|---|---|---|
| TiO2 NPs | Oxygen | 43.34 | 69.61 |
| Titanium | 56.66 | 30.39 | |
| Total | 100.00 | 100.00 |
3.3. Cell viability assay
To investigate the potency of our green TiO2 NPs drug delivery system, a cell viability assay was conducted on the breast adenocarcinoma model (MCF), and the lung cancer cell line A-549. A normal non-tumorigenic cell line, MCF10A, served as a control to evaluate the apparent safety profile and selectivity index (SI). MCF-7 and A-549 are well-established models to study the effect of antitumor drugs on breast and lung cancer cells, respectively.64–67 The half-maximal inhibitory concentration (IC50) for TiO2 NPs against MCF-7 was shown to be 1.73 μg mL−1, whereas the IC50 of TiO2 NPs against the A-549 cell line was 4.79 μg mL−1. The 50% cytotoxicity concentration of the NPs against the control cell line was 15.7 μg mL−1.The selectivity index (SI) for MCF-7 breast cancer cell lineage was a notable 8.77, indicating its pronounced therapeutic index, while SI for A-549 was a tolerable 3.17 (Table 3 and Fig. 5). A previous study was done on 4T1 breast BALB/c mouse tumor model using a different formulation of TiO2 NPs, where the IC50 was 4.11 μg mL−1.68 Sonochemically-synthesized TiO2 NPs had an IC50 of 60 μg mL−1 on MCF-7.69 A recent study conducted on a triple-negative breast cancer model using a different green TiO2 NPs formulation (propolis-extract) showed a much higher IC50 of 18.7 μg mL−1.70
| IC50 on MCF-7 (μg mL−1) | CC50 on MCF10A | SI (CC50/IC50) |
|---|---|---|
| 1.73 | 15.17 | 8.77 |
| IC50 on A549 (μg mL−1) | CC50 on MCF10A | SI (CC50/IC50) |
|---|---|---|
| 4.79 | 15.17 | 3.17 |
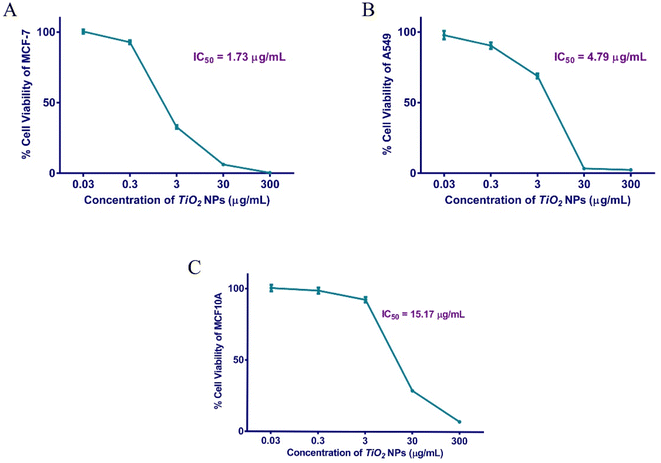 | ||
| Fig. 5 Cellular viability of human MCF-7 (A), A-549 (B), and MCF10A (C) cells after exposure to different concentrations of TiO2 NPs for 48 h. | ||
Similarly, our prepared TiO2 NPs displayed strong anti-proliferative activity against the A-549 lung cancer model as compared to a previous study.67 This cell viability assay indicates a potential candidate as a selective antineoplastic agent. Our data suggests the preferential uptake of the designed NPs by the leaky cancer cells, as compared to the normal control.71 In addition, our findings proposed the safe potential use of TiO2 NPs as a future alternative to the current chemotherapeutics that possess several systemic toxic effects.72
3.4. Apoptosis assay
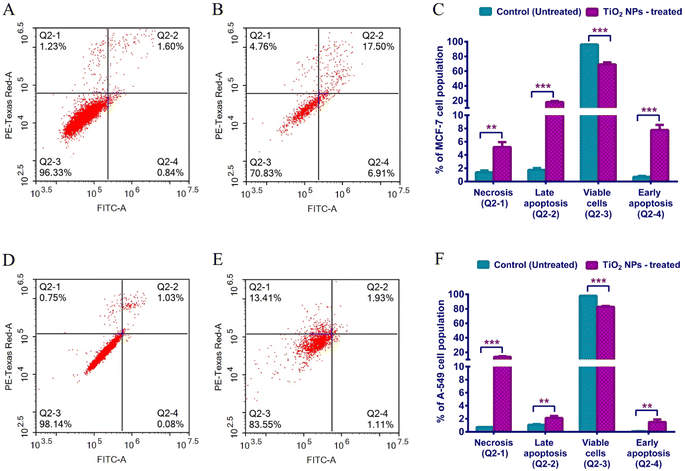
Apoptosis is the regulated cell suicide that is activated upon incurred DNA damage, infection, stress, or aberrant cellular process. It is a life-saving mechanism to prevent oncogenic transformation. Apoptosis is remarkably deregulated in cancer, displaying a hallmark of immortality and uncontrolled proliferation.73,74 Cells undergoing apoptosis shrink, show cell membranes blebbing, DNA fragmented, cytoskeleton degraded, and phagocytosis signaled.74,75 Tumor cells send various factors into the surrounding microenvironment to repress programmed cell death. In order to elucidate the molecular mechanism of our TiO2 NPs-induced cell death, a standard apoptosis assay was conducted.The gold-standard Annexin V/Propidium Iodide (AV/PI) assay was carried out to estimate the percentage of cells that have undergone programmed cell death (apoptosis) or inflammatory cell death (necrosis).75,76 Living cells maintain an intact phospholipid membrane, internalizing phosphatidyl serine and preventing the entry of charged dyes into the cells. However, when cells die by apoptosis, the plasma membrane loses its integrity, displaying phosphatidyl serine facing outward.75 Annexin V binds phosphatidyl serine and emits a signal that is detected by a Flow Cytometer. Propidium iodide intercalates the DNA of cells undergoing necrosis.77
MCF-7 cells treated for 48 h with TiO2 NPs were compared to untreated cells. Our findings revealed that 17.96% of treated MCF-7 were in the late apoptotic phase, while 5.2% were in the necrotic stage. A-549 treated similarly showed 13.64% of the population in the necrosis stage (Table 4 and Fig. 6). The apoptotic and anticancer activities of TiO2 NPs against cancer cells have been previously reported.69,78 For instance, Wang et al. displayed a significant potential for TiO2 NPs to stimulate apoptosis and produce DNA damage in A-549 cells.78
| Apoptotic stage | Percent cell populationa | |||
|---|---|---|---|---|
| MCF-7 cells | A-549 cells | |||
| Control | TiO2 treated | Control | TiO2 treated | |
| a The percent cell population is recorded as the average of triplicates ± SD. ** and *** indicatestatistical significance from the control where P-value ≤ 0.01 and ≤0.00, respectively. | ||||
| Necrosis cells (Q2-1) | 1.393 ± 0.2658 | **5.203 ± 0.7506 | 0.7267 ± 0.0208 | ***13.64 ± 0.9658 |
| Late apoptosis (Q2-2) | 1.76 ± 0.2685 | ***17.96 ± 1.517 | 1.070 ± 0.1153 | **2.133 ± 0.2937 |
| Viable cells (Q2-3) | 96.15 ± 0.168 | ***69.08 ± 2.801 | 98.13 ± 0.0907 | ***82.72 ± 1.227 |
| Early apoptosis (Q2-4) | 0.70 ± 0.1311 | ***7.76 ± 0.7758 | 0.07667 ± 0.0153 | **1.510 ± 0.3904 |
Apparently, the two cell lines responded differently after exposure to TiO2 NPs for 48 hours. MCF-7 showed a significant programmed cell suicide, remarkably in late apoptosis, relative to necrotic cell death in A-549 cells. These results indicate the strong potential of greenly-synthesized TiO2 NPs as candidate chemotherapeutic agents exerting an apoptotic effect in breast cancer.
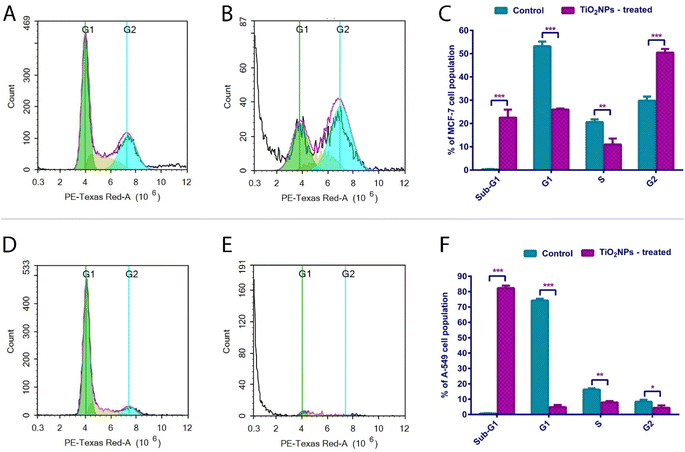
3.5. Cell cycle assay
Actively dividing cells follow precise stages known as the cell cycle. In the first phase, G1, the cell grows and accumulates nutrients, organelles, and proteins needed for DNA replication. Meanwhile, the cells duplicate their chromosomes in the S and G2 phases and produce proteins needed for division, respectively. Finally, in the M stage, mitosis, the parent cell divides equally into two daughters. On the other hand, undividing cells or those lacking the necessary nutrient/growth factors for DNA replication enter a quiescent phase. For the cell to proceed from one stage into the following, specialized proteins (cyclins and cyclin-dependent kinases) come into play. Moreover, checkpoints occur to validate the process and make sure no errors occur. G1/S and G2/M are hallmark checkpoints regulating the cell transition through the cell cycle phases. Checkpoints are precisely orchestrated by cyclins, kinases and modulator (activator/inhibitor) proteins.79 A cell cycle assay is based on quantifying DNA concentrations and visualizing chromatin patterns in different cell cycle stages. This is detected using a flow cytometer.79–81A standard cell cycle assay was conducted according to the established guidelines82,83 to interpret the cell death mechanisms of MCF-7 and A-549 cells exposed to TiO2 NPs for 48 hours relative to the untreated controls.
Interestingly, 22.52% of TiO2 NPs treated MCF-7 cells were arrested in the Sub-G1 phase. A remarkable 50.43% of TiO2 NPs treated MCF-7 cells were arrested in the G2 phase of the cell cycle before proceeding to mitotic division (Table 5 and Fig. 7). This showed a marked reduction of cellular population in G1 relative to the reference cells. Less than 11% of the cellular population was in the S phase. This aligns with our findings that a significant proportion of MCF-7 cells undergo late apoptosis. Taken together, MCF-7 cells treated with TiO2 NPs show a hallmark apoptotic profile early enough in the cell cycle. It represents a dynamic cell cycle arrest model at multiple phases, insinuating interplay with modulators of alternative checkpoints. This is particularly significant in tumors escaping one checkpoint and evading cell cycle arrest. Anticancer drugs targeting the G2 phase offer a plausible choice if tumor cells evade the G1/S checkpoint.84
| Cell cycle phase | MCF-7 | A-549 | ||
|---|---|---|---|---|
| Control (untreated) | TiO2-treated | Control (untreated) | TiO2-treated | |
| * refers to p-value ≤ 0.05 vs. control cells, ** refers to p-value ≤ 0.01 vs. control cells and *** refers to p-value ≤ 0.001 vs. control cells. | ||||
| Sub-G1 | 0.3233 ± 0.1557 | ***22.52 ± 3.455 | 0.7133 ± 0.1137 | ***82.30 ± 1.570 |
| G1 | 53.16 ± 2.061 | ***25.99 ± 0.49 | 74.17 ± 1.130 | ***4.837 ± 1.341 |
| S | 20.59 ± 1.202 | **10.98 ± 2.525 | 16.25 ± 0.8317 | **7.947 ± 0.8903 |
| G2 | 29.78 ± 1.768 | ***50.43 ± 1.595 | 8.317 ± 1.241 | *4.350 ± 1.599 |
On the other hand, 82.3% of A-549 treated cells were shown to be in the Sub-G1 phase, denoting an extensive DNA fragmentation/loss event and a pronounced ability to cease cell cycle progression before the G1/S checkpoint.85 Our findings suggest complementary evidence of the pronounced cytotoxicity of the synthesized TiO2 NPs. Our findings aligned very well with a previous study which reported a highly remarkable elevation in the frequency of several mammalian cells in the sub-G1 phase of the cell cycle following exposure to TiO2 NPs.86
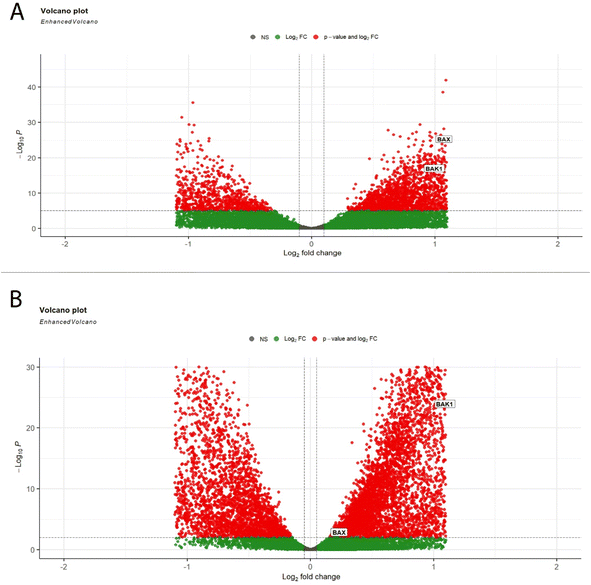
3.6. Bioinformatic analysis to select top targeted genes for RT-qPCR analysis
Our bioinformatics workflow was primarily concerned with identifying differentially expressed genes that showed differences in expression in tumor samples comparable to normal solid samples in both TCGA-BRCA and TCGA-LUSC. This difference was assessed in terms of both fold-change (FC) and statistical significance adjusted p-value. In both datasets, the volcanic plot illustrated that Bax and Bak1 genes were upregulated and differentially expressed in tumor samples compared to normal samples, as shown in Fig. 8. Based on the obtained results, Bax and Bak1 genes were selected for further gene expression analysis using RT-qPCR.3.7. Gene expression for apoptotic markers
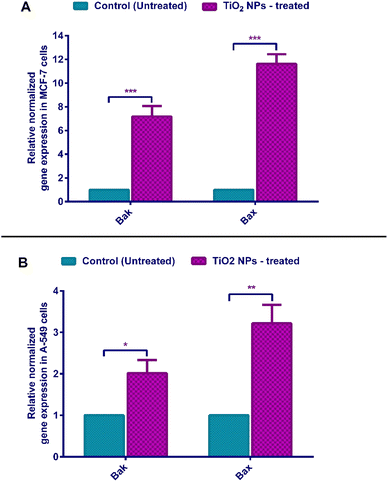
Apoptosis could be induced by either intrinsic or extrinsic pathways. The intrinsic pathway, also known as mitochondrial-mediated cell death, is activated upon the reception of an apoptotic stimulus. This is subsequently followed by activating a series of proteins, eventually leading to the oligomerization of the cytosolic Bax and Bak proteins. Then, these apoptotic proteins insert themselves into the outer mitochondrial membrane, leading to pore-formation and increased mitochondrial permeability. Afterwards, cytochrome C, is released from the mitochondria into the cytoplasm, activating an apoptosome and caspase 9.87 These mediators eventually activate effector caspases 3 and 7, leading to DNA degradation, cell membrane blebs, cytoskeleton destruction, cell shrinkage, and phagocytosis by immune cells. Cancer cells cannot usually undergo programmed-cell suicide and thus replicate indefinitely.76Quantitative PCR (qPCR) was carried out to assess the relative normalized gene expression of two apoptosis hallmark genes, namely, “Bcl-2 associated X protein” (Bax) and “Bcl-2 antagonist/killer” (Bak). β-Actin was used as the internal reference (housekeeping gene).88 Upon treatment with TiO2 NPs, MCF-7 cells show a 7.17-fold change in BakAK and 11.62-fold in Bax expression (Fig. 9). Treated A-549 cells witnessed a 2.01 and 3.21 folds increase for Bak and Bax, respectively (Fig. 9). These findings complement our apoptosis and cell cycle results, which indicate that TiO2 NPs induce programmed cell suicide through the Bax/Bak-mediated outer membrane permeabilization (MOMP),75,83,89 which successively leads to caspase-dependent apoptosis. Interestingly, Bak and Bax proteins were recently shown to orchestrate necrotic death87 observed in A-549 cells.90–92 Hence, our greenly synthesized TiO2 NPs are strong candidate antitumor agents causing neat cell death (via intrinsic apoptotic pathway).
3.8. Immunoblotting for apoptotic proteins
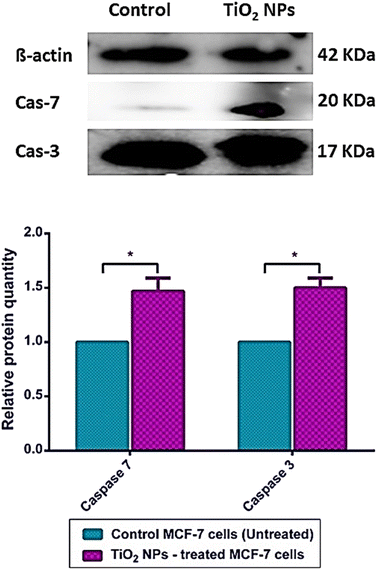
To further confirm the mechanism of action of our greenly synthesized TiO2 NPs as a potential pro-apoptotic agent, the protein expression of caspases 3 and 7 was quantifiably analyzed by western blot, and the quantification was done utilizing the ChemiDoc imaging system. Our findings showed that caspases 3 and 7 are significantly overexpressed (about 1.5 increase) in TiO2 NPs treated MCF-7 relative to untreated cells (Fig. 10). Likewise, TiO2 NPs were noticed to significantly elevate the expression of caspase-3 in the hepatoma cell line (HepG2), as well as the activity of caspase 3/7.93 In another study, the TiO2 NPs synthesized with the aid of Lactobacillus bacteria have also caused a significant upregulation of caspase 3 in human colorectal cancer cells (HT-29) and actuation of the intrinsic apoptosis pathway.94 A previous study on human hepatocarcinoma cells (SMMC-7721) revealed potentiated effects for TiO2 NPs when combined with doxorubicin (DOX) in increasing the caspase-3 protein compared to sole DOX treatment.95Meanwhile, treatment of SMMC-7721 hepatocarcinoma with TiO2 NPs alone showed a slight, not yet significant, increase in caspase 3 protein expression.95 Our findings agree with the results of the apoptosis and cell cycle assays above, which showed that TiO2 NPs induce mitochondrial-mediated apoptotic cytolysis.
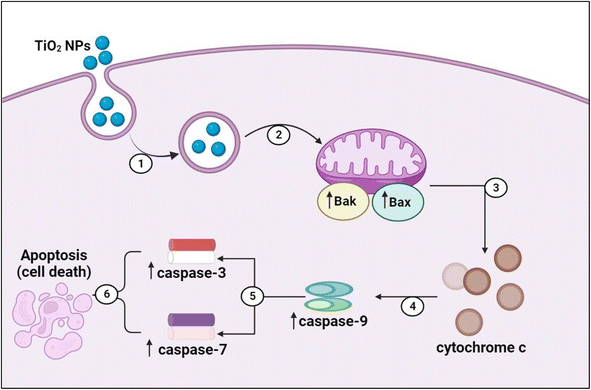
Caspases are critical proteins for regulating programmed cell death and inflammatory reactions. Thus, inadequate caspase activation can endorse carcinogenesis. In this regards, caspases have been generally classified by their established functions in programmed cell death into caspase-3, -6, -7, -8, and -9, which are sub-categorized according to their mode of action into initiator caspases (caspase-8 and -9) and killer caspases (caspase-3, -6, and -7).96 TiO2 NPs are proposed to exert their anticancer activities via mitochondrion-based cell death (Fig. 11). In this context, TiO2 NPs are internalized into cancer cells via endosomal uptake. Then, intracellularly, the TiO2 NPs significantly increased the gene expression of Bak and Bax apoptotic markers and their relative proteins. This stimulates the intrinsic apoptotic pathway where Bak and Bax are translocated into the mitochondrion of the cancer cell, increasing the permeability of its outer membrane and thus releasing cytochrome C pro-apoptotic protein from the mitochondrial matrix into the cytosol.91,92,96 The mitochondrial perturbation and cytochrome C release activate the initiator caspase 9. Once activated, caspase 9 activates the executioners of apoptosis (caspases 3 and 7) via internal cleavage of large and small sub-units. The effector caspases 3 and 7 induce apoptosis via protein crosslinking, DNA fragmentation, and hydrolysis of nuclear proteins.96
3.9. Molecular docking of TiO2 NPs in the active sites of caspase-3 and EGFR-TK
TiO2 NPs could bind to both crystal structure pockets with binding affinities of −4.16 and −4.06 kcal mol−1 for PDB IDs 2XYG and 1M17, respectively. Fig. 12A shows the TiO2 NPs bound to caspase-3 active pocket (shown in green sticks), including 7 main residues: Arg 64, Ser 120, His 121, Gly 122, Gln 161, Ala 162 and Cys 163. The NP's two oxygen atoms formed 4 H-bonds; 2 of them were with Gly 122, one with Ser 120, and the last with His 121.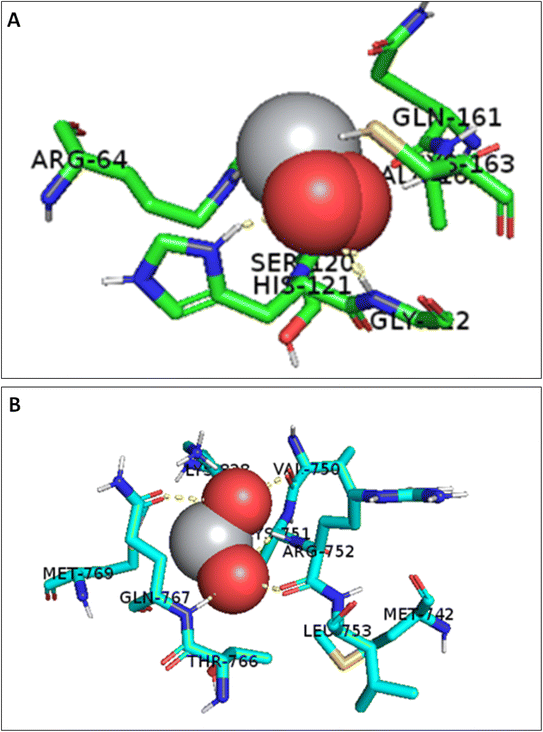 | ||
| Fig. 12 (A) 3D interactions of TiO2 NPs with caspase 3 active pocket (PDB ID: 2XYG) and (B) 3D interactions of TiO2 NPs with EGFR-TK active pocket (PDB ID: 1M17). The active site residues are shown in blue sticks. | ||
EGFR-TK is widely overexpressed in breast and lung cancers, where it contributes to the tumorigenesis process by promoting cellular growth, angiogenesis, metastasis, and resistance to chemotherapy.97 In this context, a growing body of evidence has emerged over the past 20 years regarding the targeted inhibition of the EGFR-TK family as a cancer treatment approach.97 The current study demonstrated several binding interactions for TiO2 NPs, where it was able to form 2 H-bonds with Gln-767, 2 H-bonds with Arg 752, and an H-bond with Val 750 in the active site of EGFR-TK, Fig. 12B. Therefore, TiO2 NPs is suggested to interfere with EGFR-TK and reduce cellular proliferation by limiting the activity of EGFR-TK in the tumor model. The study suggests further preclinical investigations for using TiO2 NPs as an EGFR-TK inhibitor.
4. Conclusion
There is a dire need to discover novel chemotherapeutic agents or enhance the efficacy of existing ones. To this end, we biosynthesized TiO2 NPs using an eco-friendly novel sonochemical approach and tested its effect on two cancer cell lines. We used Ocimum basilicum essential oil as a natural reductant and capping agent to efficiently biosynthesize TiO2 NPs with outstanding surface properties and biochemical profiles. Our TiO2 NPs exhibited potent cytotoxic effects on breast (MCF-7) and lung cancer (A-549) cell populations while maintaining an excellent selectivity index on normal cells. Apoptosis and cell cycle assays indicated that TiO2 NPs induced apoptosis and arrested the cell cycle in the subG1 phase in both tumor models. Gene expression analysis confirmed the up-regulation of BAX and BAK genes, emphasizing the intrinsic mitochondrial apoptosis mechanism. Protein expression analysis of caspases 3 and 7 enzymes showed a remarkable over-expression relative to the control untreated cells. The molecular docking study also demonstrated good binding potential for TiO2 NPs with two of the common targets in breast and lung cancers (caspase 3 and EGFR-TK). In a nutshell, we herein report the green synthesis of TiO2 NPs with potent cytotoxicity and high selectivity profiles capable of inducing apoptosis in both breast and lung cancer cells via the mitochondrial-mediated permeability approach. Moreover, the TiO2 NPs exhibited several binding interactions with EGFR-TK, reinforcing their ability to induce apoptosis and inhibit cellular growth.Data availability
Data is contained within the article and ESI.†Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was funded by RSC Research Fund grant (ID: R22-3870733873) from the Royal Society of Chemistry to Dr Sherif Ashraf Fahmy. The authors are thankful to the Deanship of Scientific Research at King Khalid University for funding this work through general research project with project grant number GRP/111/44. Also, the authors thank Nanogate-Egypt Company for conducting the Transmission Electron Microscopy (TEM) analysis and taking the TEM images for the samples in the company's facility.References
- NCI, Cancer Statistics, 2023, https://www.cancer.gov/about-cancer/understanding/statistics Search PubMed.
- WHO, W.H.O. Cancer, 2023, https://www.who.int/health-topics/cancer#tab=tab_1 Search PubMed.
- D. Hanahan and R. A. Weinberg, Hallmarks of cancer: the next generation, Cell, 2011, 144(5), 646–674 CrossRef CAS PubMed.
- F. Bray, et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2018, 68(6), 394–424 CrossRef PubMed.
- T. A. Moo, et al., Overview of Breast Cancer Therapy, PET Clin., 2018, 13(3), 339–354 CrossRef PubMed.
- B. Mansoori, et al., The Different Mechanisms of Cancer Drug Resistance: A Brief Review, Adv. Pharm. Bull., 2017, 7(3), 339–348 CrossRef CAS PubMed.
- I. Ritacco, et al., Hydrolysis in acidic environment and degradation of satraplatin: a joint experimental and theoretical investigation, Inorg. Chem., 2017, 56(10), 6013–6026 CrossRef CAS PubMed.
- G. Mattheolabakis and C. M. Mikelis, Nanoparticle Delivery and Tumor Vascular Normalization: The Chicken or The Egg?, Front. Oncol., 2019, 9, 1227 CrossRef PubMed.
- A. H. AbuBakr, H. A. Hassan, A. Abdalla, O. M. Khowessah and G. A. Abdelbary, Therapeutic potential of cationic bilosomes in the treatment of carrageenan-induced rat arthritis via fluticasone propionate gel, Int. J. Pharm., 2023, 635, 122776 CrossRef CAS PubMed.
- S. A. Fahmy, et al., Ozonated Olive Oil: Enhanced Cutaneous Delivery via Niosomal Nanovesicles for Melanoma Treatment, Antioxidants, 2022, 11(7), 1318 CrossRef CAS PubMed.
- R. A. Youness, et al., Oral Delivery of Psoralidin by Mucoadhesive Surface-Modified Bilosomes Showed Boosted Apoptotic and Necrotic Effects against Breast and Lung Cancer Cells, Polymers, 2023, 15(6), 1464 CrossRef CAS PubMed.
- S. A. Fahmy, et al., Molecular Engines, Therapeutic Targets, and Challenges in Pediatric Brain Tumors: A Special Emphasis on Hydrogen Sulfide and RNA-Based Nano-Delivery, Cancers, 2022, 14(21), 5244, DOI:10.3390/cancers14215244.
- N. K. Sedky, et al., Box–Behnken design of thermo-responsive nano-liposomes loaded with a platinum (IV) anticancer complex: evaluation of cytotoxicity and apoptotic pathways in triple negative breast cancer cells, Nanoscale Adv., 2023, 5, 5399–5413 RSC.
- N. K. Sedky, et al., Co-Delivery of Ylang Ylang Oil of Cananga odorata and Oxaliplatin Using Intelligent pH-Sensitive Lipid-Based Nanovesicles for the Effective Treatment of Triple-Negative Breast Cancer, Int. J. Mol. Sci., 2023, 24(9), 8392 CrossRef CAS PubMed.
- R. Khursheed, et al., Biomedical applications of metallic nanoparticles in cancer: Current status and future perspectives, Biomed. Pharmacother., 2022, 150, 112951 CrossRef CAS PubMed.
- A. R. Al Jayoush, H. A. Hassan, H. Asiri, M. Jafar, R. Saeed, R. Harati and M. Haider, Niosomes for nose-to-brain delivery: a non-invasive versatile carrier system for drug delivery in neurodegenerative diseases, J. Drug Delivery Sci. Technol., 2023, 105007 CrossRef CAS.
- S. G. Abonashey, H. A. Hassan, M. A. Shalaby, A. G. Fouad, E. Mobarez and H. A. El-Banna, Formulation, pharmacokinetics, and antibacterial activity of florfenicol-loaded niosome, Drug Delivery Transl. Res., 2023, 1–16 Search PubMed.
- M. H. Alyami, et al., Retama monosperma chemical profile, green synthesis of silver nanoparticles, and antimicrobial potential: a study supported by network pharmacology and molecular docking, RSC Adv., 2023, 13(37), 26213–26228 RSC.
- P. S. Jassal, et al., Green synthesis of titanium dioxide nanoparticles: Development and applications, J. Agric. Food Res., 2022, 100361 Search PubMed.
- C. Zarzzeka, et al., Use of titanium dioxide nanoparticles for cancer treatment: A comprehensive review and bibliometric analysis, Biocatal. Agric. Biotechnol., 2023, 102710 CrossRef CAS.
- H. Iqbal, et al., Breast cancer inhibition by biosynthesized titanium dioxide nanoparticles is comparable to free doxorubicin but appeared safer in BALB/c mice, Materials, 2021, 14(12), 3155 CrossRef CAS PubMed.
- R. Mund, et al., Novel titanium oxide nanoparticles for effective delivery of paclitaxel to human breast cancer cells, J. Nanopart. Res., 2014, 16, 1–12 CrossRef CAS.
- S. Sharmin, et al., Nanoparticles as antimicrobial and antiviral agents: A literature-based perspective study, Heliyon, 2021, 7(3), e06456 CrossRef CAS PubMed.
- S. A. Fahmy, et al., Green synthesis of platinum and palladium nanoparticles using Peganum harmala L. seed alkaloids: Biological and computational studies, Nanomaterials, 2021, 11(4), 965 CrossRef CAS PubMed.
- S. Ying, et al., Green synthesis of nanoparticles: Current developments and limitations, Environ. Technol. Innovation, 2022, 26, 102336 CrossRef CAS.
- M. Aravind, M. Amalanathan and M. S. M. Mary, Synthesis of TiO2 nanoparticles by chemical and green synthesis methods and their multifaceted properties, SN Appl. Sci., 2021, 3, 1–10 Search PubMed.
- M. Altikatoglu, et al., Green synthesis of copper oxide nanoparticles using Ocimum basilicum extract and their antibacterial activity, Fresenius Environ. Bull., 2017, 25(12), 7832–7837 Search PubMed.
- B. Thakur, A. Kumar and D. Kumar, Green synthesis of titanium dioxide nanoparticles using Azadirachta indica leaf extract and evaluation of their antibacterial activity, S. Afr. J. Bot., 2019, 124, 223–227 CrossRef CAS.
- N. M. Aboeita, et al., Enhanced anticancer activity of nedaplatin loaded onto copper nanoparticles synthesized using red algae, Pharmaceutics, 2022, 14(2), 418 CrossRef CAS PubMed.
- H. A. Salam and R. Sivaraj, Ocimum basilicum L. var. purpurascens Benth.-LAMIACEAE Mediated Green Synthesis and Characterization of Titanium Dioxide Nanoparticles, Adv. Bio Res., 2014, 5(3), 10–16 Search PubMed.
- A. R. Malik, et al., Green synthesis of RGO-ZnO mediated Ocimum basilicum leaves extract nanocomposite for antioxidant, antibacterial, antidiabetic and photocatalytic activity, J. Saudi Chem. Soc., 2022, 26(2), 101438 CrossRef CAS.
- A. S. Abdelsattar, et al., Utilization of Ocimum basilicum extracts for zinc oxide nanoparticles synthesis and their antibacterial activity after a novel combination with phage, Mater. Lett., 2022, 309, 131344 CrossRef CAS.
- K. Arshad Qamar, et al., Anticancer activity of Ocimum basilicum and the effect of ursolic acid on the cytoskeleton of MCF-7 human breast cancer cells, Lett. Drug Des. Discovery, 2010, 7(10), 726–736 CrossRef.
- W. Tan, et al., Surface coating changes the physiological and biochemical impacts of nano-TiO2 in basil (Ocimum basilicum) plants, Environ. Pollut., 2017, 222, 64–72 CrossRef CAS PubMed.
- N. Lagopati, et al., Nanomedicine: Photo-activated nanostructured titanium dioxide, as a promising anticancer agent, Pharmacol. Ther., 2021, 222, 107795 CrossRef CAS PubMed.
- X. Zhu, K. Pathakoti and H.-M. Hwang, Green synthesis of titanium dioxide and zinc oxide nanoparticles and their usage for antimicrobial applications and environmental remediation, in Green Synthesis, Characterization and Applications of Nanoparticles, Elsevier, 2019, pp. 223–263 Search PubMed.
- S. A. Fahmy, et al., Green extraction of essential oils from Pistacia lentiscus resins: encapsulation into Niosomes showed improved preferential cytotoxic and apoptotic effects against breast and ovarian cancer cells, J. Drug Delivery Sci. Technol., 2023, 87, 104820 CrossRef CAS.
- N. K. Mahdy, et al., Toward Scaling up the Production of Metal Oxide Nanoparticles for Application on Washable Antimicrobial Cotton Fabrics, ACS Omega, 2022, 7(43), 38942–38956 CrossRef CAS PubMed.
- S. Kamiloglu, et al., Guidelines for cell viability assays, Food Front., 2020, 1(3), 332–349 CrossRef.
- T. D. Schmittgen and K. J. Livak, Analyzing real-time PCR data by the comparative CT method, Nat. Protoc., 2008, 3(6), 1101–1108 CrossRef CAS PubMed.
- C. E. Bohl, et al., Structural basis for accommodation of nonsteroidal ligands in the androgen receptor, J. Biol. Chem., 2005, 280(45), 37747–37754 CrossRef CAS PubMed.
- P. Wee and Z. Wang, Epidermal growth factor receptor cell proliferation signaling pathways, Cancers, 2017, 9(5), 52 CrossRef PubMed.
- M. Westwood, et al., Epidermal growth factor receptor tyrosine kinase (EGFR-TK) mutation testing in adults with locally advanced or metastatic non-small cell lung cancer: a systematic review and cost-effectiveness analysis, Health Technol. Assess., 2014, 18(32), 1 Search PubMed.
- S. Chibber and I. Ahmad, Molecular docking, a tool to determine interaction of CuO and TiO2 nanoparticles with human serum albumin, Biochem. Biophys. Rep., 2016, 6, 63–67 Search PubMed.
- L. Jin, et al., Breast cancer lung metastasis: Molecular biology and therapeutic implications, Cancer Biol. Ther., 2018, 19(10), 858–868 CrossRef CAS PubMed.
- M. Z. Ahmad, et al., Green synthesis of titanium dioxide nanoparticles using ocimum sanctum leaf extract: in vitro characterization and its healing efficacy in diabetic wounds, Molecules, 2022, 27(22), 7712 CrossRef CAS PubMed.
- V. Malapermal, et al., Enhancing antidiabetic and antimicrobial performance of Ocimum basilicum, and Ocimum sanctum (L.) using silver nanoparticles, Saudi J. Biol. Sci., 2017, 24(6), 1294–1305 CrossRef CAS PubMed.
- P. R. More, et al., Antimicrobial applications of green synthesized bimetallic nanoparticles from Ocimum basilicum, Pharmaceutics, 2022, 14(11), 2457 CrossRef CAS PubMed.
- D. P. Bezerra, et al., The dual antioxidant/prooxidant effect of eugenol and its action in cancer development and treatment, Nutrients, 2017, 9(12), 1367 CrossRef PubMed.
- É. R. Q. Dos Santos, et al., Linalool as a therapeutic and medicinal tool in depression treatment: a review, Curr. Neuropharmacol., 2022, 20(6), 1073 CrossRef PubMed.
- A. F. Ahmed, et al., Antioxidant activity and total phenolic content of essential oils and extracts of sweet basil (Ocimum basilicum L.) plants, Food Sci. Hum. Wellness, 2019, 8(3), 299–305 CrossRef.
- T. Aburjai, et al., Chemical Compositions and Anticancer Potential of Essential Oil from Greenhouse-cultivated Ocimum basilicum Leaves, Indian J. Pharm. Sci., 2020, 82(1), 178–183 Search PubMed.
- O. V. Kharissova, et al., Greener synthesis of chemical compounds and materials, R. Soc. Open Sci., 2019, 6(11), 191378 CrossRef PubMed.
- J. O. Tijani, M. M. Ndamitso and A. S. Abdulkareem, Facile synthesis and characterization of TiO2 nanoparticles: X-ray peak profile analysis using Williamson–Hall and Debye–Scherrer methods, Int. Nano Lett., 2021, 11, 241–261 CrossRef.
- N. Horti, et al., Synthesis and photoluminescence properties of titanium oxide (TiO2) nanoparticles: Effect of calcination temperature, Optik, 2019, 194, 163070 CrossRef CAS.
- R. Aswini, S. Murugesan and K. Kannan, Bio-engineered TiO2 nanoparticles using Ledebouria revoluta extract: Larvicidal, histopathological, antibacterial and anticancer activity, Int. J. Environ. Anal. Chem., 2021, 101(15), 2926–2936 CrossRef CAS.
- W. Ahmad, K. K. Jaiswal and S. Soni, Green synthesis of titanium dioxide (TiO2) nanoparticles by using Mentha arvensis leaves extract and its antimicrobial properties, Inorg. Nano-Met. Chem., 2020, 50(10), 1032–1038 CrossRef CAS.
- T. Luttrell, et al., Why is anatase a better photocatalyst than rutile?-Model studies on epitaxial TiO2 films, Sci. Rep., 2014, 4(1), 4043 CrossRef PubMed.
- O. Alameer, et al., Expoloriting of graphene oxide for improving physical properties of TiO2 (NPs): toward photovoltaic devices and wastewater remediation approaches, Eur. Phys. J. Plus, 2022, 137(10), 1160 CrossRef CAS.
- N. Raval, et al., Importance of physicochemical characterization of nanoparticles in pharmaceutical product development, Basic Fundam. Drug Delivery, 2019, 369–400 CAS.
- U. Baig, et al., Single step production of high-purity copper oxide-titanium dioxide nanocomposites and their effective antibacterial and anti-biofilm activity against drug-resistant bacteria, Mater. Sci. Eng., C, 2020, 113, 110992 CrossRef CAS PubMed.
- M. A. E. A. A. A. El-Remaily, A. M. Abu-Dief and O. Elhady, Green synthesis of TiO2 nanoparticles as an efficient heterogeneous catalyst with high reusability for synthesis of 1,2-dihydroquinoline derivatives, Appl. Organomet. Chem., 2019, 33(8), e5005 CrossRef.
- S. A. Fahmy, et al., Thermosensitive Liposomes Encapsulating Nedaplatin and Picoplatin Demonstrate Enhanced Cytotoxicity against Breast Cancer Cells, ACS Omega, 2022, 7(46), 42115–42125 CrossRef CAS PubMed.
- H.-r. Moon, et al., Subtype-specific characterization of breast cancer invasion using a microfluidic tumor platform, PLoS One, 2020, 15(6), e0234012 CrossRef CAS PubMed.
- L. Ziko, et al., Antibacterial and anticancer activities of orphan biosynthetic gene clusters from Atlantis II Red Sea brine pool, Microb. Cell Factories, 2019, 18(1), 1–16 CrossRef PubMed.
- K. Nohara, F. Wang and S. Spiegel, Glycosphingolipid composition of MDA-MB-231 and MCF-7 human breast cancer cell lines, Breast Cancer Res. Treat., 1998, 48, 149–157 CrossRef CAS PubMed.
- L. Armand, et al., Long-term exposure of A549 cells to titanium dioxide nanoparticles induces DNA damage and sensitizes cells towards genotoxic agents, Nanotoxicology, 2016, 10(7), 913–923 CrossRef CAS PubMed.
- S. Abdel-Ghany, et al., Vorinostat-loaded titanium oxide nanoparticles (anatase) induce G2/M cell cycle arrest in breast cancer cells via PALB2 upregulation, 3 Biotech, 2020, 10(9), 407 CrossRef PubMed.
- K. Murugan, et al., Hydrothermal synthesis of titanium dioxide nanoparticles: mosquitocidal potential and anticancer activity on human breast cancer cells (MCF-7), Parasitol. Res., 2016, 115(3), 1085–1096 CrossRef PubMed.
- M. T. Hamed, et al., Novel synthesis of titanium oxide nanoparticles: Biological activity and acute toxicity study, Bioinorg. Chem. Appl., 2021, 2021, 8171786 Search PubMed.
- S. A. Fahmy, et al., Synthesis, characterization and host-guest complexation of asplatin: improved in vitro cytotoxicity and biocompatibility as compared to cisplatin, Pharmaceuticals, 2022, 15(2), 259 CrossRef CAS PubMed.
- I. Ritacco, et al., Hydrolysis in acidic environment and degradation of satraplatin: a joint experimental and theoretical investigation, Inorg. Chem., 2017, 56(10), 6013–6026 CrossRef CAS PubMed.
- A. R. Safa, Drug and apoptosis resistance in cancer stem cells: a puzzle with many pieces, Cancer Drug Resist., 2022, 5(4), 850 CrossRef CAS PubMed.
- C. M. Neophytou, et al., Apoptosis deregulation and the development of cancer multi-drug resistance, Cancers, 2021, 13(17), 4363 CrossRef CAS PubMed.
- M. Hassan, et al., Apoptosis and molecular targeting therapy in cancer, BioMed Res. Int., 2014, 2014, 150845 Search PubMed.
- C. M. Pfeffer and A. T. Singh, Apoptosis: a target for anticancer therapy, Int. J. Mol. Sci., 2018, 19(2), 448 CrossRef PubMed.
- T. Bezabeh, et al., Detection of drug-induced apoptosis and necrosis in human cervical carcinoma cells using 1H NMR spectroscopy, Cell Death Differ., 2001, 8(3), 219–224 CrossRef CAS PubMed.
- Y. Wang, et al., Cytotoxicity, DNA damage, and apoptosis induced by titanium dioxide nanoparticles in human non-small cell lung cancer A549 cells, Environ. Sci. Pollut. Res., 2015, 22, 5519–5530 CrossRef CAS PubMed.
- W. Stallaert, et al., The structure of the human cell cycle, Cell Syst., 2022, 13(3), 230–240 CrossRef CAS PubMed .e3..
- Y. Sun, et al., The influence of cell cycle regulation on chemotherapy, Int. J. Mol. Sci., 2021, 22(13), 6923 CrossRef CAS PubMed.
- A. E. Eastman and S. Guo, The palette of techniques for cell cycle analysis, FEBS Lett., 2020, 594(13), 2084–2098 CrossRef CAS PubMed.
- G. I. Shapiro and J. W. Harper, Anticancer drug targets: cell cycle and checkpoint control, J. Clin. Invest., 1999, 104(12), 1645–1653 CrossRef CAS PubMed.
- D. Plesca, S. Mazumder and A. Almasan, DNA damage response and apoptosis, Methods Enzymol., 2008, 446, 107–122 CAS.
- T. Kawabe, G2 checkpoint abrogators as anticancer drugs, Mol. Cancer Ther., 2004, 3(4), 513–519 CrossRef CAS PubMed.
- B.-Y. Peng, et al., AGA induces sub-G1 cell cycle arrest and apoptosis in human colon cancer cells through p53-independent/p53-dependent pathway, BMC Cancer, 2023, 23(1), 1–13 CrossRef CAS PubMed.
- H. Chang, et al., Effect of Titanium Dioxide Nanoparticles on Mammalian Cell Cycle In Vitro: A Systematic Review and Meta-Analysis, Chem. Res. Toxicol., 2022, 35(9), 1435–1456 Search PubMed.
- J. Karch, et al., Bax and Bak function as the outer membrane component of the mitochondrial permeability pore in regulating necrotic cell death in mice, eLife, 2013, 2, e00772 Search PubMed.
- M. Wang and M. Thanou, Targeting nanoparticles to cancer, Pharmacol. Res., 2010, 62(2), 90–99 CrossRef CAS PubMed.
- G. Dewson and R. M. Kluck, Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis, J. Cell Sci., 2009, 122(16), 2801–2808 CrossRef CAS PubMed.
- C. Garrido, et al., Mechanisms of cytochrome c release from mitochondria, Cell Death Differ., 2006, 13(9), 1423–1433 CrossRef CAS PubMed.
- A. Peña-Blanco and A. J. García-Sáez, Bax, Bak and beyond—mitochondrial performance in apoptosis, FEBS J., 2018, 285(3), 416–431 CrossRef PubMed.
- T. T. Renault and J. E. Chipuk, Death upon a kiss: mitochondrial outer membrane composition and organelle communication govern sensitivity to BAK/BAX-dependent apoptosis, Chem. Biol., 2014, 21(1), 114–123 CrossRef CAS PubMed.
- Z. Xia, et al., Titanium dioxide nanoparticles induce mitochondria-associated apoptosis in HepG2 cells, RSC Adv., 2018, 8(55), 31764–31776 RSC.
- R. Vigneshwaran, D. Ezhilarasan and S. Rajeshkumar, Inorganic titanium dioxide nanoparticles induces cytotoxicity in colon cancer cells, Inorg. Chem. Commun., 2021, 133, 108920 CrossRef CAS.
- Y. Chen, et al., Anticancer efficacy enhancement and attenuation of side effects of doxorubicin with titanium dioxide nanoparticles, Int. J. Nanomed., 2011, 2321–2326 CAS.
- S. A. Lakhani, et al., Caspases 3 and 7: key mediators of mitochondrial events of apoptosis, Science, 2006, 311(5762), 847–851 CrossRef CAS PubMed.
- M. Mubeen and S. G. Kini, A review on the design and development of EGFR tyrosine kinase inhibitors in cancer therapy, Int. J. Ther. Appl., 2012, 5, 29–37 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra08908h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |