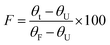Open Access Article
Open Access ArticleImproving the thermostability and modulating the inulin profile of inulosucrase through rational glycine-to-proline substitution†
Thanapon Charoenwongpaiboon *a,
Nawapat Sommanata,
Karan Wangpaiboonbc,
Manatsanan Puangpathanachaib,
Piamsook Pongsawasdib and
Rath Pichyangkurab
*a,
Nawapat Sommanata,
Karan Wangpaiboonbc,
Manatsanan Puangpathanachaib,
Piamsook Pongsawasdib and
Rath Pichyangkurab
aDepartment of Chemistry, Faculty of Science, Silpakorn University, Nakhon Pathom, 73000, Thailand. E-mail: charoenwongpaib_t@su.ac.th
bDepartment of Biochemistry, Faculty of Science, Chulalongkorn University, Bangkok, 10330, Thailand
cCenter of Excellence in Structural and Computational Biology, Department of Biochemistry, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
First published on 11th January 2024
Abstract
The flexibility of protein structure plays a crucial role in enzyme stability and catalysis. Among the amino acids, glycine is particularly important in conferring flexibility to proteins. In this study, the effects of flexible glycine residues in Lactobacillus reuteri 121 inulosucrase (LrInu) on stability and inulin profile were investigated through glycine-to-proline substitutions. Molecular dynamics (MD) simulations were employed to discover the flexible glycine residues, and eight glycine residues, including Gly217, Gly298, Gly330, Gly416, Gly450, Gly624, Gly627, Gly629, were selected for site-directed mutagenesis. The results demonstrated significant changes in both thermostability and inulin profiles of the variants. Particularly, the G624P and G627P variants showed reduced production of long-chain oligosaccharides compared to the WT. This can be ascribed to the increased rigidity of the active site, which is crucial for the induction-fit mechanism. Overall, this study provides valuable insights into the role of flexible glycine residues in the activity, stability, and inulin synthesis of LrInu.
1. Introduction
Flexibility of protein structure is important for enzyme stability and catalysis. It plays a role in substrate binding, allowing enzymes to adapt their active sites and bind substrates with specificity.2 Flexible structure enables enzymes to undergo conformational changes that stabilize the transition state, lowering the activation energy and promoting efficient reactions. Moreover, flexibility allows enzymes to adapt to different environmental conditions and maintain their activity. It also plays a role in allosteric regulation,3,4 where conformational changes conducted from regulatory sites affect enzyme activity. Therefore, flexibility is crucial for enzymes as it enables substrate recognition, efficient catalysis, stability, and regulatory mechanisms.The glycine residue is unique among the amino acids because it has a hydrogen atom as a side chain. It has greater conformational freedom, allowing it to provide flexibility for adjacent residues. Numerous studies have revealed that glycine plays a special role in enzyme structure and function. The mutation of specific glycine residues may decrease the stability and activity of these enzymes.5 On the contrary, the proline residue is mostly steric compared to other amino acids since its side chain is connected to the amino group, forming a pyrrolidine loop. Therefore, substituting certain flexible residues with proline can possibly enhance the protein's thermostability.6–8 The glycine-to-proline mutation has been utilized to enhance the thermostability of methyl parathion hydrolase (MPH) from Ochrobactrum sp. M231, resulting in an increase in both the melting temperature and operation time.9 Substitution of proline for Gly244 of esterase from Psychrobacter sp. could increase half-life (t1/2) of the enzyme at 40 °C from 16 min to 11.6 h.10 In addition, our recent study reveal that G249P mutations of Bacillus licheniformis levansucrase significantly increase both the enzyme's stability and levan yield.11
Inulosucrase is an enzyme that is present in various bacteria, such as Leuconostoc citreum,12 Lactobacillus reuteri,13 Lactobacillus johnsonii,14 and Streptomyces viridochromogenes.15 This enzyme exhibits transfructosylation activity, allowing it to convert sucrose into inulin. Inulin is a soluble dietary fiber consisting of fructose molecules linked by a β-2,1 glycosidic bond. Inulin was not digested by the human gastrointestinal tract, making it an ideal prebiotic that supports the growth of profitable bacteria in the gut.16 Additionally, inulin has a mildly sweet taste, making it a potential sugar substitute in processed foods.17 It is also categorized as a low-glycemic index sweetener, indicating that it has minimal impact on blood sugar levels.18 Notably, bacterial inulosucrase is capable of synthesizing higher molecular weight inulin compared to the inulin found in plants.19
Previous studies have successfully employed rational enzyme engineering of inulosucrase to control the size of the resulting product20 and to enhance its stability.21 Furthermore, specific amino acid residues that potentially play a role in substrate binding have been identified in previous research.22 Nevertheless, the role of glycine residues located in the flexible regions of inulosucrase remains unexplored. In this study, we investigated the impact of flexible glycine residues in Lactobacillus reuteri 121 inulosucrase (LrInu) through glycine-to-proline substitutions. Initially, molecular dynamics simulations and Root Mean Square Deviation (RMSD) analysis were employed to identify the flexible regions of LrInu. Subsequently, the flexible glycine residues were replaced with proline to enhance structural rigidity. Thermostability and obtained inulin profiles of wild-type (WT) and variant LrInu were then examined. The findings revealed significant changes in thermostability and inulin profiles for certain glycine-to-proline mutants.
2. Experimental
2.1 Construction, expression, and purification of LrInu
Site-directed mutagenesis was carried out using the PCR overlap extension technique,23 with oligonucleotide primers specified in Table S1.† PrimeStar™ DNA polymerase (Takara Bio, Japan) was utilized for DNA amplification. The PCR product obtained inserted into the pET-21b vector via XhoI and NdeI restriction sites. This combined construct was introduced into Escherichia coli Top10 cells. These recombinant cells were placed onto LB agar enriched with 100 μg mL−1 of ampicillin and then kept at 37 °C. A colony was cultivated in LB broth, containing 100 μg mL−1 ampicillin, to produce the plasmid. The resultant recombinant plasmid was subsequently confirmed through sequencing. The recombinant plasmid was transformed into E. coli BL21 (DE3). The LrInus were expressed and purified according to the protocol described in a previous study.12.2 Enzyme activity assay
Sucrase activity was evaluated using the DNS assay.24 The purified enzymes were incubated with 500 μl of 250 mM sucrose solution containing a 50 mM sodium acetate buffer (pH 5.5) and 1 mM CaCl2 at 50 °C. Subsequently, 500 μl of DNS reagent were added to the reaction and boiled for 10 min. The quantity of reducing sugars produced by the enzymes was determined at 540 nm using a spectrophotometer. One unit of sucrase activity was defined as the enzyme amount required to release 1 μmol of glucose per min. To identify the optimum temperature for catalytic activity of LrInus, an activity assay was conducted at pH 5.5 at different temperatures.2.3 Thermostability analysis
The half-life of both the WT and variant LrInu enzymes was evaluated using a similar method as described previously.1 In brief, the enzymes (0.02 mg mL−1) were incubated in a 50 mM sodium acetate buffer (pH 5.5) at 50 °C for a duration of 0–8 h. Subsequently, the remaining enzyme activity were assessed using DNS assay at various time intervals. To calculate the half-life values, the data obtained was plotted as remaining activity versus time curves and fitted with an exponential decay equation.T50 values were determined by incubating the purified enzymes (0.02 mg mL−1) in a 50 mM sodium acetate buffer (pH 5.5) over a range of temperatures from 32 to 80 °C for 60 min. Subsequently, the residual activity of each enzyme variant was assessed at a temperature of 50 °C. Graphs depicting the relationship between activity and temperature were generated and then fitted with the Boltzmann sigmoid equation. The minimum and maximum activity values obtained from the Boltzmann sigmoid equation were designated as the values corresponding to the unfolded protein (θU) and the folded protein (θF). The fraction folded (F) was computed using the equation:
2.4 Inulin synthesis and analysis
Inulosucrase (2 U mL−1) was incubated with 250 mM sucrose in the presence of 50 mM acetate buffer (pH 5.5) and 1 mM CaCl2. The mixtures were then incubated at 30 °C for 24 h. To terminate the reaction, the mixtures were boiled for 10 min. The sugar composition in the reaction mixture was analyzed using high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD), as previously described.25 To determine the mass of oligosaccharide products, the MALDI-TOF mass spectroscopy technique was employed utilizing the JEOL™ SpiralTOF MALDI Imaging-TOF/TOF Mass Spectrometer (JMS-S3000). The matrix used for this analysis was 2,5-dihydroxybenzoic acid (DHB).2.5 Computational methods
The homology models of the LrInu were generated from crystal structure of Lactobacillus johnsonii inulosucrase (PDB ID: 2YFS with 74.17% identity) using the SWISS-MODEL server.26 The protonation state of all amino acids in the models was determined at pH 5.5 using the H++ server.27 MD simulations were conducted at 323 K and 400 K using the AMBER20 package. The models were immersed in a truncated octahedral box filled with the TIP3P water model, maintaining a buffer distance of 13 Å, utilizing the AMBER20 LEaP module. The systems were then neutralized by adding sodium ions (Na+).Subsequently, minimization was executed to eliminate unfavorable interactions through a series of procedures, each consisting of 2500 steepest-descent steps and 2500 conjugated gradient steps, in which the heavy atoms of proteins were initially restrained with the force constant of 5.0 kcal mol−1 Å−2, followed by the restraint of protein backbones using force constants of 10, 5, and 1 kcal mol−1 Å−2. Finally, all of the systems were minimized without any restraining force, the PMEMD module of AMBER20 was employed to perform system simulations under the previously mentioned condition.11 The Langevin dynamic technique28 was employed to control temperatures, with a collision frequency of 1 ps⁻1. During the heating stage, the system underwent a temperature increase from 0 to 323 K or 400 K over 200 ps, while restraining protein backbones with a force constant of 10 kcal mol⁻1 Å⁻2 in the NVT ensemble. Additionally, the systems underwent a 300 ps equilibration stage at 323 K or 400 K in the NVT ensemble. The production stage involved simulations at 323 K or 400 K and 1 atm in the NPT ensemble for 100 ns, generating a total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 conformations for each independent run. This entire process was conducted in two replicates for each system. The Cpptraj module of AMBER20 (ref. 29) was utilized for calculation of Root Mean Square Deviation (RMSD), Root-Mean-Square Fluctuation (RMSF) and hydrogen bond (H-bond). Note that the RMSF values were computed based on the alpha carbon and beta carbon atoms (or hydrogen atoms for glycine) of each residue. The presence of hydrogen bonds between the protein and ligands was assessed based on specific criteria: (i) the distance between the H-bond donor (HD) and H-bond acceptor (HA) ≤ 3.0 Å and (ii) the angle between the donor–H–acceptor (HD–H⋯HA) of ≥135°.11 Furthermore, the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method was applied using the MMPBSA.py module to calculate the total binding free energy (ΔGbind).30
000 conformations for each independent run. This entire process was conducted in two replicates for each system. The Cpptraj module of AMBER20 (ref. 29) was utilized for calculation of Root Mean Square Deviation (RMSD), Root-Mean-Square Fluctuation (RMSF) and hydrogen bond (H-bond). Note that the RMSF values were computed based on the alpha carbon and beta carbon atoms (or hydrogen atoms for glycine) of each residue. The presence of hydrogen bonds between the protein and ligands was assessed based on specific criteria: (i) the distance between the H-bond donor (HD) and H-bond acceptor (HA) ≤ 3.0 Å and (ii) the angle between the donor–H–acceptor (HD–H⋯HA) of ≥135°.11 Furthermore, the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method was applied using the MMPBSA.py module to calculate the total binding free energy (ΔGbind).30
3. Results
3.1 Identification of highly flexible glycine using molecular dynamics simulation
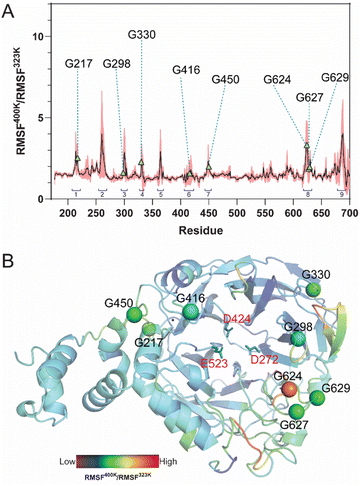
MD simulations of LrInu model were performed at 323 K and 400 K for 100 ns. The stability of the system was assessed using the Root-Mean-Square Deviation (RMSD) analysis. High structural fluctuation of LrInu was observed during the first 60 ns and then eventually found equilibrium (Fig. S1†). Therefore, the MD trajectories from 80 to 100 ns were extracted for further Root-Mean-Square Fluctuation (RMSF) analysis. As shown in Fig. 1, RMSF400K/RMSF323K ratio revealed nine highly flexible regions of the LrInu that respond to the change in temperature. Most of these regions have glycine residues. Due to the fact that glycine, unlike other amino acids, features a hydrogen atom as its side chain, resulting in significantly greater conformational flexibility within the structure. Therefore, the glycine residues in flexible regions, including Gly217, Gly298, Gly330, Gly416, Gly450, Gly624, Gly627 and Gly629, were substituted by Pro with the aim that they might increase thermostability of LrInu.3.2 Specific activity and optimum temperature of variant enzymes
The candidate variants were constructed, expressed, and purified. The activity assay demonstrated that most of the variants showed a significant decrease in sucrase activity compared to the WT, except for G416P (Fig. 2A). The G217P, G330P, G450P, G624P, G627P, and G629P variants exhibited relative activities of 63%, 62%, 23%, 24%, 51%, and 58%, respectively. Additionally, the activity of G298P was not detectable, suggesting the critical role of this residue in LrInu's catalysis. After that, the effect of temperature on the catalytic activity of WT and variants was evaluated at 25-70 °C (Fig. 2B). The results showed that all variants and the WT had similar optimum temperatures of 50 °C and lost almost all activity at 60 °C.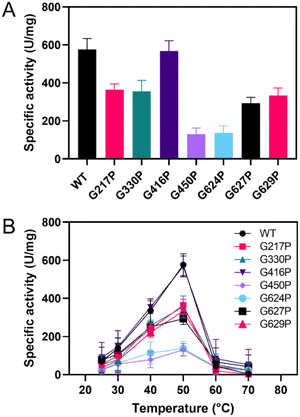 | ||
| Fig. 2 (A) Specific activity of WT and variant LrInu at 50 °C. (B) Effect of temperature on activity of WT and variants at 25–70 °C. | ||
3.3 Thermostability of variant enzymes
Thermostability parameters of the WT LrInu and its variants were evaluated (Fig. 3). The kinetic stability of all variants was investigated at WT's optimal temperature of 50 °C. As shown in Fig. 3A, G416P, G624P, G627P, and G629P exhibited higher sucrase activity after an 8 hour incubation compared to that of the WT,1 resulting in a 1.5-fold, 1.4-fold, 2.2-fold, and 1.6-fold increase in the T1/2 value, respectively, as compared to the WT (Table 1). To assess the impact of mutations on LrInu's thermodynamic stability, T50 of the enzymes were measured. The T50 value, the midpoint of the enzyme inactivation, of G416P, G624P, G627P, and G629P was greater than that of the WT (Table 1), indicating that certain mutations can enhance thermodynamic stability of LrInu.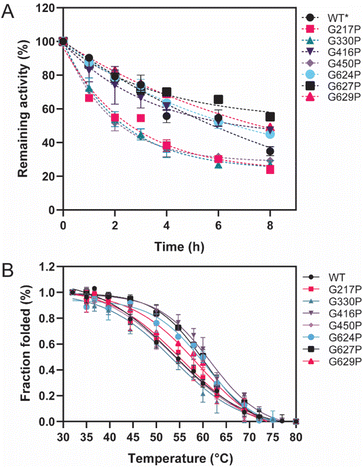 | ||
| Fig. 3 Thermostability of WT and variant LrInu. (A) Kinetic stability of WT and variant LrInu evaluated at 50 °C. (B) Melting temperature analysis of WT and variant LrInu determined by thermal inactivation assay. *Data from ref. 1. | ||
3.4 Product profile
HPAEC-PAD and MALDI-TOF spectroscopy were employed to investigate the product profile and size distribution of fructooligosaccharides (FOS) synthesized by both the WT and variant LrInu enzymes, as presented in Fig. 4. The HPAEC chromatogram demonstrated that the mutations did not have an impact on the linkage within oligosaccharides, as all observed peaks on the chromatogram were comparable (Fig. 4A). However, MALDI-TOF analysis revealed significant differences in the degree of polymerization (DP) between the mutant inulin and the WT inulin (Fig. 4B). Specifically, G416P, G450P, and G624P variants generated shorter inulin FOS compared to the WT, while the G627P variant produced longer-chain FOS. For G217P, G330P, and G629P variants, although they were able to produce FOS with a similar DP as the WT, the HPAEC intensity indicated that these mutants generated a lower quantity of DP7, and longer-chain FOS compared to the WT. These findings suggest that the flexibility of LrInu not only affects its stability but also its processivity.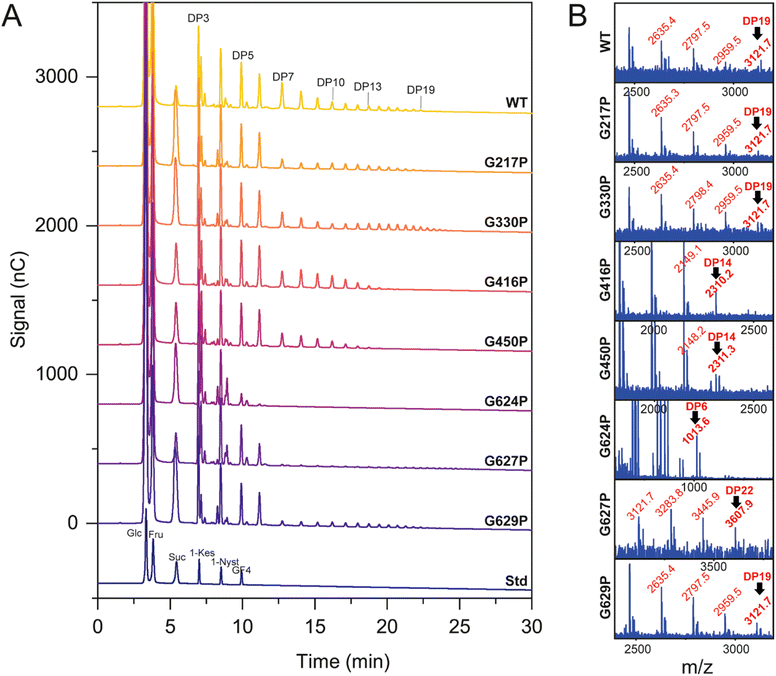 | ||
| Fig. 4 Product profile of WT and variant LrInu analyzed by (A) HPAEC-PAD and (B) MALDI-TOF mass spectroscopy. | ||
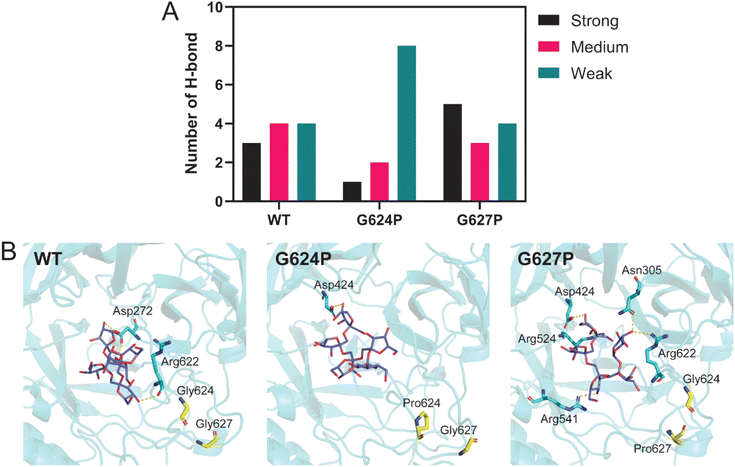
3.5 Binding of FOS on WT, G624P and G627P LrInu
Although located in the same loop, both G624P and G627P variants display different product profiles. To investigate the impact of these mutations, molecular dynamics (MD) simulations and MM/GBSA analysis were performed. The systems, including WT, G624P, and G627P variants in complex with 1-β-fructofuranosyl-nystose (GF4), were simulated at 323 K for 100 ns. The MD trajectories from 80 to 100 ns were extracted for further H-bond and MM/GBSA analyses. As shown in Table 2, the binding affinity of G624P was relatively lower than that of WT, while G627P exhibited the highest affinity among them. This finding supports the MALDI-TOF result that G627P can produce a higher DP. Since proline has a cyclic structure and a limited phi angle, proline substitution may change the conformation of nearby residues, impacting the interaction between the enzyme and substrate. H-bond analysis revealed that proline substitution affects the number of H-bonds between LrInu and FOS (GF4) (Fig. 5A and B). The number of strong H-bonds increases for G627P, supporting the reason why this variant produces higher DP FOS. In contrast, the G624P mutation decreased the number of strong and medium H-bonds between the enzyme and FOS.| Energy component (kcal mol−1) | WT | G624P | G627P |
|---|---|---|---|
| Gas term | |||
| ΔEvdW | −54.7 ± 0.6 | −57.1 ± 0.7 | −57.6 ± 0.4 |
| ΔEele | −106.6 ± 1.9 | −113.5 ± 1.8 | −99.1 ± 1.2 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Solvent term | |||
| ΔGpoalr | 130.4 ± 1.3 | 142.0 ± 1.4 | 123.5 ± 0.9 |
| ΔGnon-polar | −8.4 ± 0.1 | −8.7 ± 0.1 | −9.1 ± 0.0 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Binding free energy | |||
| ΔGbind (MM/GBSA) | −39.3 ± 1.0 | −37.3 ± 0.9 | −42.3 ± 0.6 |
4. Discussion
The glycine residue possesses a unique characteristic among amino acids, as its side chain consists of hydrogen atom. This distinctive feature allows for greater conformational freedom, thereby providing flexibility to adjacent residues. Given this property, it is not surprising that glycine plays a significant role in the structure and function of enzymes.2 In this study, molecular dynamics simulations were employed to identify the highly flexible glycine residues in LrInu. Based on the conservation analysis (Fig. S2†), it was observed that most of the flexible glycine residues, with the exception of Gly624, were highly conserved among inulosucrases, suggesting their potential significance in their catalysis or stability. To further examine this point, a rigid amino acid proline was used to substitute the flexible glycine residues of LrInu. The rationale behind this substitution was to reduce the conformational degrees of freedom of the enzyme, thus possibly altering LrInu's stability and product profile.As a result, mutations in flexible glycine residues have an impact on both the activity and stability of LrInu. Specifically, the activity of the G298P variant could not be detected, likely due to its close proximity to the active site. Moreover, G217P, G330P and G450P variants exhibit much lower activity and stability compared to WT. Notably, Gly450 is positioned on the loop that is connected to the calcium binding site of LrInu, which has been previously reported to play a crucial role in enzyme activity and stability.22,31 The impact of the G330P mutation on LrInu activity is relatively lower compared to that of G450P, as Gly330 is positioned further away from the active site and the oligosaccharide binding site of LrInu. However, it is important to highlight that this residue is highly conserved among inulosucrases, suggesting its essential role in maintaining the native structure of LrInu.
Other mutants including G416P, G624P, G627P and G629P were found to be more stable compared to WT, although they have lower activity than that of WT (except G416P). It is well known that substitutions with proline residues decrease the conformational degrees of freedom in the polypeptide chain thus enhancing the stability of the protein at higher temperatures.32 This approach has been successfully employed in various enzymes, such as α-glucosidase,33 lipase,8 endoglucanase,7 levansucrase11 and transaminase.6 According to the RMSF analysis, G624P, G627P, and G629P are located within the loop, which is highly sensitive to temperature changes. Introducing the rigid amino acid proline into this loop is expected to decrease its flexibility and subsequently enhance protein stability. Nevertheless, it is noteworthy that this loop connects to the active site of LrInu, which provides an explanation for the strong decrease in enzyme activity. Surprisingly, the G416P mutation had no effect on enzyme activity, possibly because it was located away from the active site.
In addition to affecting enzyme activity and stability, proline substitutions also have an impact on the oligosaccharide profile of LrInu. This can be attributed to the increased rigidity of the active site, which is a crucial feature for the induction-fit mechanism.2,34 Consequently, most variants produce shorter-chain or lower amounts of long-chain oligosaccharides compared to the WT. Furthermore, the model indicates that Gly416 and Gly450 are situated on the loop connecting to the calcium binding site of the enzyme, which has been previously identified as playing a significant role in product elongation.22 Interestingly, the G627P variant produces longer-chain inulin than WT, potentially due to the higher rigidity of the active site, which enhance the interaction to the acceptor molecule. This hypothesis was validated through MD simulation. RMSF analysis distinctly illustrated that the G627P mutation could decrease the flexibility of LrInu's amino acid residues (Fig. S3†). Certain regions, specifically residues 256–266, 301–305, 332–336, 415–417, 547–552, and 627–629, situated on the binding pocket's rim, indicated their potential significance in enzyme catalysis and stability. Overall, this study has provided valuable data regarding the impact of flexible glycine residues on LrInu's activity, stability, and inulin synthesis.
5. Conclusions
This study investigates the role of flexible glycine residues in the activity, stability, and inulin synthesis of Lactobacillus reuteri 121 inulosucrase (LrInu). The substitution of the rigid amino acid proline for these flexible glycine residues can result in both an increase and a decrease in the stability of LrInu. Additionally, mutations have varying effects on enzyme activity and the obtained product profile, depending on their position. Specifically, the G416P, G624P, G627P, and G629P variants were found to exhibit higher stability compared to the WT, suggesting their potential for further development in inulin synthesis. Overall, this study addresses a research gap in the field of inulosucrase and inulin biosynthesis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work (Grant No. RGNS 64-215) was supported by Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (OPS MHESI), Thailand Science Research and Innovation (TSRI) and Silpakorn University, Thailand. K. W. would like to acknowledge the support from Chulalongkorn University (CE66_036_2300_008).References
- T. Charoenwongpaiboon, K. Wangpaiboon, M. Puangpathanachai, P. Pongsawasdi and R. Pichyangkura, Appl. Microbiol. Biotechnol., 2023, 107, 6831–6843 CrossRef CAS PubMed.
- B. X. Yan and Y. Q. Sun, J. Biol. Chem., 1997, 272, 3190–3194 CrossRef CAS PubMed.
- X. Zhu, M. Byrnes, J. W. Nelson and S. H. Chang, Biochemistry, 1995, 34, 2560–2565 CrossRef CAS PubMed.
- C. R. Meyer, J. A. Bork, S. Nadler, J. Yirsa and J. Preiss, Arch. Biochem. Biophys., 1998, 353, 152–159 CrossRef CAS PubMed.
- Y. Feng, S. Huang, W. Zhang, Z. Zeng, X. Zou, L. Zhong, J. Peng and G. Jing, Biochimie, 2004, 86, 893–901 CrossRef CAS PubMed.
- Z. Xie, L. Zhai, D. Meng, Q. Tian, Z. Guan, Y. Cai and X. Liao, 3 Biotech, 2020, 10, 323 CrossRef PubMed.
- A. S. Dotsenko, S. Pramanik, A. V. Gusakov, A. M. Rozhkova, I. N. Zorov, A. P. Sinitsyn, M. D. Davari and U. Schwaneberg, Biochem. Eng. J., 2019, 152, 107395 CrossRef CAS.
- D. Yuan, Z. Zhao, X. Wang, S. Guo, B. Yang and Y. Wang, Eur. J. Lipid Sci. Technol., 2016, 118, 821–826 CrossRef CAS.
- J. Tian, P. Wang, S. Gao, X. Chu, N. Wu and Y. Fan, FEBS J., 2010, 277, 4901–4908 CrossRef CAS PubMed.
- L. Kulakova, A. Galkin, T. Nakayama, T. Nishino and N. Esaki, Biochim. Biophys. Acta, 2004, 1696, 59–65 CrossRef CAS PubMed.
- M. Klaewkla, K. Wangpaiboon, R. Pichyangkura and T. Charoenwongpaiboon, Proteins: Struct., Funct., Bioinf., 2023, 92(2), 170–178 CrossRef PubMed.
- M. E. Ortiz-Soto, V. Olivares-Illana and A. López-Munguía, Biocatal. Biotransform., 2004, 22, 275–281 CrossRef CAS.
- S. A. F. T. v. Hijum, G. H. v. Geel-Schutten, H. Rahaoui, M. J. E. C. v. d. Maarel and L. Dijkhuizen, Appl. Environ. Microbiol., 2002, 68, 4390–4398 CrossRef PubMed.
- M. A. Anwar, S. Kralj, M. J. E. C. v. d. Maarel and L. Dijkhuizen, Appl. Environ. Microbiol., 2008, 74, 3426–3433 CrossRef CAS PubMed.
- H.-J. Frasch, S. S. v. Leeuwen and L. Dijkhuizen, Microbiology, 2017, 163, 1030–1041 CrossRef CAS PubMed.
- S. Macfarlane, G. T. Macfarlane and J. H. Cummings, Aliment. Pharmacol. Ther., 2006, 24, 701–714 CrossRef CAS PubMed.
- M. J. Mabel, P. T. Sangeetha, K. Platel, K. Srinivasan and S. G. Prapulla, Carbohydr. Res., 2008, 343, 56–66 CrossRef CAS PubMed.
- J. L. Causey, J. M. Feirtag, D. D. Gallaher, B. C. Tungland and J. L. Slavin, Nutr. Res., 2000, 20, 191–201 CrossRef CAS.
- D. C. Mudannayake, D. D. Jayasena, K. M. S. Wimalasiri, C. S. Ranadheera and S. Ajlouni, Int. J. Food Sci. Technol., 2022, 57, 5764–5780 CrossRef CAS.
- T. Charoenwongpaiboon, T. Sitthiyotha, P. P. Na Ayutthaya, K. Wangpaiboon, S. Chunsrivirot, M. Hengsakul Prousoontorn and R. Pichyangkura, Carbohydr. Polym., 2019, 209, 111–121 CrossRef CAS PubMed.
- D. Ni, S. Zhang, O. Kırtel, W. Xu, Q. Chen, E. T. Öner and W. Mu, J. Agric. Food Chem., 2021, 69, 13125–13134 CrossRef CAS PubMed.
- T. Charoenwongpaiboon, P. Punnatin, M. Klaewkla, P. Pramoj Na Ayutthaya, K. Wangpaiboon, S. Chunsrivirot, R. A. Field and R. Pichyangkura, ACS Omega, 2020, 5, 28001–28011 CrossRef CAS PubMed.
- K. L. Heckman and L. R. Pease, Nat. Protoc., 2007, 2, 924–932 CrossRef CAS PubMed.
- G. L. Miller, Anal. Chem., 1959, 31, 426–428 CrossRef CAS.
- T. Charoenwongpaiboon, K. Wangpaiboon, M. Klaewkla, K. Kuttiyawong, R. A. Field and R. Pichyangkura, Biochem. Eng. J., 2022, 185, 108524 CrossRef CAS.
- A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. T. Heer, T. A. P. de Beer, C. Rempfer, L. Bordoli, R. Lepore and T. Schwede, Nucleic Acids Res., 2018, 46, W296–W303 CrossRef CAS PubMed.
- J. C. Gordon, J. B. Myers, T. Folta, V. Shoja, L. S. Heath and A. Onufriev, Nucleic Acids Res., 2005, 33, W368–W371 CrossRef CAS PubMed.
- X. Wu and B. R. Brooks, Chem. Phys. Lett., 2003, 381, 512–518 CrossRef CAS.
- D. R. Roe and T. E. Cheatham III, J. Chem. Theory Comput., 2013, 9, 3084–3095 CrossRef CAS PubMed.
- S. Genheden and U. Ryde, Expet Opin. Drug Discov., 2015, 10, 449–461 CrossRef CAS PubMed.
- L. K. Ozimek, G. J. W. Euverink, M. J. E. C. van der Maarel and L. Dijkhuizen, FEBS Lett., 2005, 579, 1124–1128 CrossRef CAS PubMed.
- K. Watanabe and Y. Suzuki, J. Mol. Catal. B: Enzym., 1998, 4, 167–180 CrossRef CAS.
- E. H. Muslin, S. E. Clark and C. A. Henson, Protein Eng. Des. Sel., 2002, 15, 29–33 CrossRef CAS PubMed.
- K. Teilum, J. G. Olsen and B. B. Kragelund, Biochim. Biophys. Acta, 2011, 1814, 969–976 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra06896j |
| This journal is © The Royal Society of Chemistry 2024 |