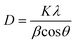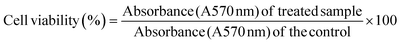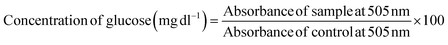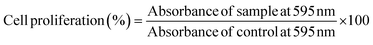Open Access Article
Open Access ArticleUnveiling the cytotoxic and anti-proliferative potential of green-synthesized silver nanoparticles mediated by Colletotrichum gloeosporioides†
Priyamvada Gupta‡
 a,
Swati Singh‡
a,
Swati Singh‡ a,
Nilesh Rai
a,
Nilesh Rai a,
Ashish Verma
a,
Ashish Verma a,
Harshita Tiwari
a,
Harshita Tiwari a,
Swapnil C. Kamble
a,
Swapnil C. Kamble b,
Hemant Kumar Gautam
b,
Hemant Kumar Gautam c and
Vibhav Gautam
c and
Vibhav Gautam *a
*a
aCentre of Experimental Medicine and Surgery, Institute of Medical Sciences, Banaras Hindu University, Varanasi-221005, India. E-mail: vibhav.gautam4@bhu.ac.in; priyamvada.24@bhu.ac.in; swati.7672@bhu.ac.in; nilesh.rai17@bhu.ac.in; ashish.4@bhu.ac.in; harshitaruchi4218@gmail.com; Tel: +918860182113
bDepartment of Technology, Savitribai Phule Pune University, Ganeshkhind, Pune 411007, India. E-mail: sckamble@unipune.ac.in
cDepartment of Immunology and Infectious Disease Biology, CSIR-Institute of Genomics and Integrative Biology, Sukhdev Vihar, New Delhi 110025, India. E-mail: hemant@igib.res.in
First published on 30th January 2024
Abstract
Fungal endophytes are a putative source of bioactive metabolites that have found significant applications in nanomedicine due to their metabolic versatility. In the present study, an aqueous extract of the fungal endophyte, Colletotrichum gloeosporioides associated with a medicinal plant Oroxylum indicum, has been used for the fabrication of green silver nanoparticles (CgAgNPs) and further evaluated their cytotoxic and anti-proliferative activity. Bioanalytical techniques including UV-Vis spectral analysis revealed a sharp band at 435 nm and functional molecules from the aqueous extract involved in the synthesis of CgAgNPs were evidenced through FTIR. Further, the crystalline nature of CgAgNPs was determined through XRD analysis and microscopy techniques including AFM, TEM and FESEM demonstrated the spherical shape of CgAgNPs exhibiting a crystalline hexagonal lattice and the size was found to be in the range of 9–29 nm. The significant cytotoxic potential of CgAgNPs was observed against breast cancer cells, MDA-MB-231 and MCF-7 with IC50 values of 18.398 ± 0.376 and 38.587 ± 1.828 μg mL−1, respectively. The biochemical study revealed that the treatment of MDA-MB-231 and MCF-7 cells with CgAgNPs reduces glucose uptake, suppresses cell proliferation, and enhances LDH release, indicating reduced cell viability and progression. Moreover, our research revealed differential expression of genes associated with apoptosis, cell cycle inhibition and metastasis suppression, evidencing anti-proliferative activity of CgAgNPs. The main objective of the present study is to harness anti-breast cancer activity of novel biogenic nanoparticles synthesized using the aqueous extract of O. indicum associated C. gloeosporioides and study the underlying mechanistic pathway exerted by these mycogenic nanoparticles.
1. Introduction
Over the past centuries, nanobiotechnology has remarkably served as a groundbreaking field in various sectors, including disease biology, engineering, chemical biology and more particularly, biomedicine.1 Recently, nanomaterials-based theranostic approach which includes diagnosis and therapeutics, has surpassed the barriers faced in eradicating the cancerous cells.2,3 These nanomaterials have inherent properties of microbicidal, anti-inflammatory, anticancer and antioxidant which have been extensively helpful in the manipulation of these nanoparticles in nanomedicine.4 The screening of these nanomaterials in in vitro and in vivo biological systems have been achieved using different microscopy-based platforms and has greatly contributed in health and medicine.5 The green-based synthesis of silver nanoparticles using microorganisms as biosynthetic machinery for the formulation of nanomaterials has gained enormous interest due to their eco-friendly, cost-effective and non-toxic nature and has also aided against various health ailments.6 Fungal endophytes associated with medicinal plants are prolific manufacturers of various bioactive compounds serving as antioxidant, antimicrobial, anti-diabetic, immunosuppressant and neuroprotective agents.7–11 Moreover, these bioactive compounds derived from fungal endophytes also exhibit potential anticancer activities against several malignancies and thus lead to the discovery of many novel compounds.12–15 The chemistry of these fungal endophyte derived compounds can be studied through several mass-spectrometry based methods coupled with bioinformatics tools so as to derive a descriptive account of their compositional structure and functional interpretation.16,17 The pharmacologically important fungal compounds have also found their applications in nanotechnology and have been used for the development of nanodrugs against cancer.18 The increased surface to volume percentage of silver suggests a better non-toxic, cost effective and eco-friendly approach for the fungal mediated formation of silver nanoparticles.19 Thus, these fungi are considered as nano-factories for mycofabrication of nanomaterials with controlled shape and size.20,21 Plethora of evidences have suggested the potential role of myco-derived silver nanoparticles against inhibition of cancer proliferation, invasion and metastasis.22–24Cancer, an unrestricted cell proliferation is considered as one of the leading and deadliest cause worldwide. Among all type of cancer, breast cancer is considered as the most common and second greatest cause of death after lung carcinoma. The most dangerous and aggressive type of cancer is triple negative breast cancer (TNBC) due to its poor prognosis, limited available treatments and metastatic property.25 The tumor proliferation rate in most of the breast cancer type is directly proportional to the expression level of the estrogen receptor. Thus, MCF-7 cell which is non-invasive type of cancer cell contains functional epidermal growth factor (EGF) and estrogen receptors whereas, MDA-MB-231 cells are more aggressive hormone independent invasive type of TNBC.26 According to the American Cancer Society, the breast cancer status reported globally among women has shown an estimate of 281![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550 and 287
550 and 287![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850 new cases of breast cancer in 2021 and 2022, respectively, showing an approximate 6300 rise and about 43
850 new cases of breast cancer in 2021 and 2022, respectively, showing an approximate 6300 rise and about 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 and 43
600 and 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 estimated deaths in 2021 and 2022, respectively that is predicted to happen in the United States.27,28 The process of alterations in metabolism and bioenergetics of tumor cells called metabolic reprogramming is considered as one of the cancer hallmarks.29 These prevalent changes in metabolism is essential for cellular growth and division of cancer cells which is required for malignant transformation, tumor invasion, unlimited proliferation, metastasis and enhanced survival capacity.30 Previous literature showed that the depletion of glucose uptake by breast cancer cells have shown increased cytotoxicity therefore, significantly reduces proliferation rate in a dose dependent manner.31 In a report, it has been demonstrated that glucose deprivation induces apoptosis in MCF-7/ADR breast cancer cells.32 The activated status and enhanced expression profile of LDH in various cancers has contributed to explore diverse characteristic features of cancer malignancies. Abnormal upregulation of LDH leads to tumor progression through high lactate production, increased glycolysis, regulating ROS production and modulating various cancer associated proteins.33 LDH is a cytosolic soluble enzyme that is released extracellularly upon impairment of cellular membrane. Therefore, its level in blood or serum could act as a signal of tissue damage.34 Previous study has shown cytotoxic potential of AgNPs on human hepatoma cancer cells that significantly exhibited rise in LDH level.35 Cell proliferation is a well distinct regulated process, critically essential in medical sector ranging from organogenesis in embryo to tissue repairment to carcinogenesis.30 Recent reports have shown the anti-proliferative action of AgNPs on HeLa cells that exhibited maximum reduction of cell proliferation.36 Deregulated cell proliferation and genomic alterations in malignant cells that abrogate normal signaling pathways governing cell cycle checkpoints, further facilitates the dysregulation of cell cycle.37 Apoptosis, a major regulator of tumor progression is governed by oncogenes and tumor suppressor gene. Previous study revealed that biogenic AgNPs induces dysregulation in gene expression of pro-apoptotic and anti-apoptotic genes against HCT-116 colon cancer cells due to nuclear damage and disruption of the mitochondrial membrane.38 Thus, the dysregulation in gene expression related to apoptosis could be considered as significant diagnostic markers in chemotherapy.39 Many reports have demonstrated the role of fungal endophytes against breast cancer progression and thus have been emerged as an effective therapeutic approach over other treatment strategies.40–42 Our previous findings showed ethyl acetate extract of fungal endophyte C. gloeosporioides isolated from O. indicum exhibits unique biochemical profile and potential cytotoxic activity.43 Considering these findings, we have selected the fungal endophyte C. gloeosporioides for the synthesis of silver nanoparticles (CgAgNPs) and assessment of their cytotoxic and anti-proliferative properties against human breast cancer cells, MDA-MB-231 and MCF-7. Several bioanalytical methods such as UV-Vis, FTIR, XRD, AFM, TEM, FESEM and EDX have been used to characterize and validate the synthesis of CgAgNPs and results revealed their fcc lattice and size ranging between 9–29 nm. Further validation of in vitro cytotoxic and anti-proliferative potential of synthesized CgAgNPs was done using MTT assay followed by biochemical assays such as glucose estimation assay, cell proliferation assay and LDH assay. The expression level of several apoptosis-related genes revealed the molecular basis of anti-proliferative activity of CgAgNPs. The novel study shows first ever synthesis of silver nanoparticles mediated by O. indicum associated fungal endophyte C. gloeosporioides and assessment of the antiproliferative activity of green nanoparticles against breast cancer cells. The significant anti-breast cancer activity of CgAgNPs further urge to explore their anti-breast cancer mechanistic study through molecular elucidation so as to establish the novel CgAgNPs as a promising biotherapeutic agent.
250 estimated deaths in 2021 and 2022, respectively that is predicted to happen in the United States.27,28 The process of alterations in metabolism and bioenergetics of tumor cells called metabolic reprogramming is considered as one of the cancer hallmarks.29 These prevalent changes in metabolism is essential for cellular growth and division of cancer cells which is required for malignant transformation, tumor invasion, unlimited proliferation, metastasis and enhanced survival capacity.30 Previous literature showed that the depletion of glucose uptake by breast cancer cells have shown increased cytotoxicity therefore, significantly reduces proliferation rate in a dose dependent manner.31 In a report, it has been demonstrated that glucose deprivation induces apoptosis in MCF-7/ADR breast cancer cells.32 The activated status and enhanced expression profile of LDH in various cancers has contributed to explore diverse characteristic features of cancer malignancies. Abnormal upregulation of LDH leads to tumor progression through high lactate production, increased glycolysis, regulating ROS production and modulating various cancer associated proteins.33 LDH is a cytosolic soluble enzyme that is released extracellularly upon impairment of cellular membrane. Therefore, its level in blood or serum could act as a signal of tissue damage.34 Previous study has shown cytotoxic potential of AgNPs on human hepatoma cancer cells that significantly exhibited rise in LDH level.35 Cell proliferation is a well distinct regulated process, critically essential in medical sector ranging from organogenesis in embryo to tissue repairment to carcinogenesis.30 Recent reports have shown the anti-proliferative action of AgNPs on HeLa cells that exhibited maximum reduction of cell proliferation.36 Deregulated cell proliferation and genomic alterations in malignant cells that abrogate normal signaling pathways governing cell cycle checkpoints, further facilitates the dysregulation of cell cycle.37 Apoptosis, a major regulator of tumor progression is governed by oncogenes and tumor suppressor gene. Previous study revealed that biogenic AgNPs induces dysregulation in gene expression of pro-apoptotic and anti-apoptotic genes against HCT-116 colon cancer cells due to nuclear damage and disruption of the mitochondrial membrane.38 Thus, the dysregulation in gene expression related to apoptosis could be considered as significant diagnostic markers in chemotherapy.39 Many reports have demonstrated the role of fungal endophytes against breast cancer progression and thus have been emerged as an effective therapeutic approach over other treatment strategies.40–42 Our previous findings showed ethyl acetate extract of fungal endophyte C. gloeosporioides isolated from O. indicum exhibits unique biochemical profile and potential cytotoxic activity.43 Considering these findings, we have selected the fungal endophyte C. gloeosporioides for the synthesis of silver nanoparticles (CgAgNPs) and assessment of their cytotoxic and anti-proliferative properties against human breast cancer cells, MDA-MB-231 and MCF-7. Several bioanalytical methods such as UV-Vis, FTIR, XRD, AFM, TEM, FESEM and EDX have been used to characterize and validate the synthesis of CgAgNPs and results revealed their fcc lattice and size ranging between 9–29 nm. Further validation of in vitro cytotoxic and anti-proliferative potential of synthesized CgAgNPs was done using MTT assay followed by biochemical assays such as glucose estimation assay, cell proliferation assay and LDH assay. The expression level of several apoptosis-related genes revealed the molecular basis of anti-proliferative activity of CgAgNPs. The novel study shows first ever synthesis of silver nanoparticles mediated by O. indicum associated fungal endophyte C. gloeosporioides and assessment of the antiproliferative activity of green nanoparticles against breast cancer cells. The significant anti-breast cancer activity of CgAgNPs further urge to explore their anti-breast cancer mechanistic study through molecular elucidation so as to establish the novel CgAgNPs as a promising biotherapeutic agent.
2. Experimental procedure
2.1. Culturing and extraction of aqueous extract of C. gloeosporioides
The fungal strain, C. gloeosporioides was cultured in 100 mL of sterile PDB (Potato Dextrose Broth) media and fermentation was done at 26 ± 2 °C, for a week in an orbital shaker incubator (100 rpm). The mycelial biomass was filtered using double layered cheesecloth and was rinsed in sterilized double distilled water (SDDW) to eliminate the traces of media content from the harvested mycelia. The collected fungal biomass (10 g) was added with 100 mL DDW for aqueous extraction and kept in an orbital shaker (100 rpm), at 28 ± 4 °C, for 24 hours. Finally, the aqueous filtrate, rich in fungal bioactive components was collected using Whatman No.1 filter paper and centrifuged to pellet down any suspended debris. The residual fungal mass was discarded and the supernatant was retained for AgNPs synthesis.2.2. Mycosynthesis of AgNPs using fungal aqueous extract
For AgNPs synthesis, 1 mM silver nitrate (AgNO3) was added to fungal aqueous extract and agitated on magnetic stirrer (350 rpm) in dark at 40 ± 4 °C for 24 hours. It was then centrifuged twice or thrice at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes following washing with nuclease free water. The supernatant was discarded and the retained pellet containing the AgNPs was lyophilized and crushed to a fine powder. Subsequently, the powdered form of AgNPs was used for characterization and in vitro experiments.
000 rpm for 15 minutes following washing with nuclease free water. The supernatant was discarded and the retained pellet containing the AgNPs was lyophilized and crushed to a fine powder. Subsequently, the powdered form of AgNPs was used for characterization and in vitro experiments.
2.3. Characterization of mycosynthesized silver nanoparticles (CgAgNPs)
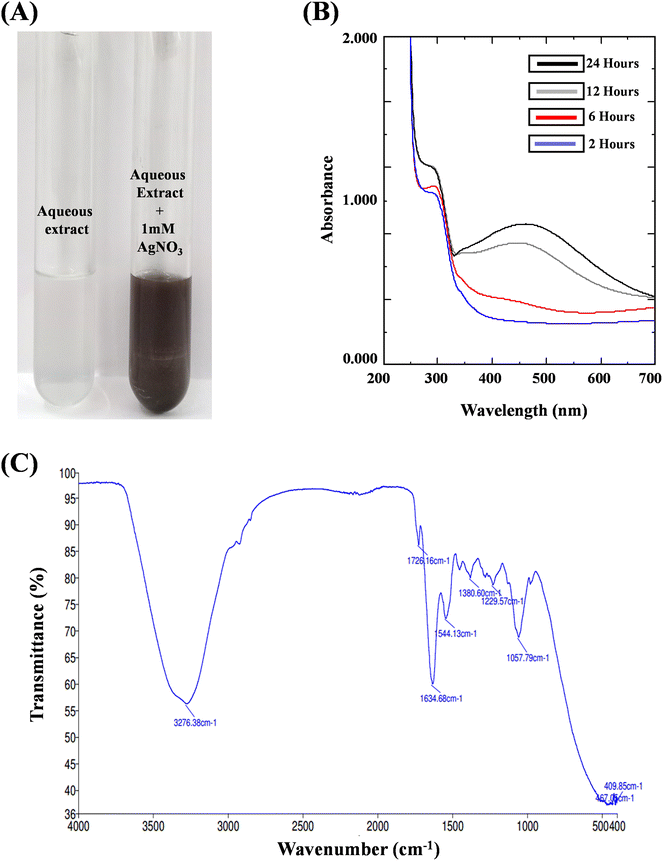
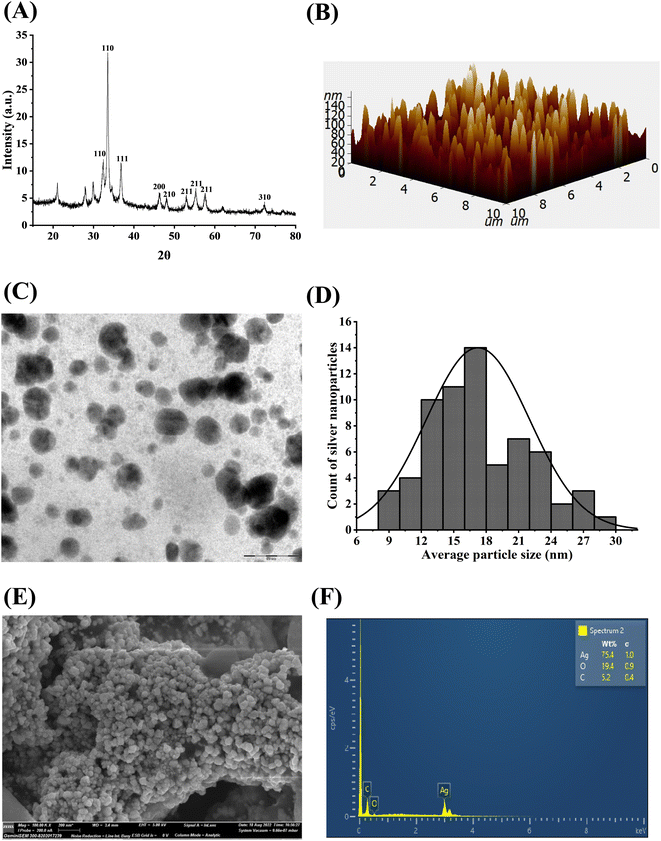
The formation of AgNPs and their optical and morphological detection were investigated through characterization using different techniques including Ultraviolet-visible spectroscopy (UV-Vis), Fourier Transform Infrared Spectroscopy (FTIR), X-ray Diffraction (XRD), Atomic Force Microscopy (AFM), Transmission Electron Microscopy (TEM) and Field Emission Scanning Electron Microscopy (FESEM).where, D = crystallite domain size, K = Scherrer constant; θ = diffraction angle, β = peak width at half of its maximum height and λ = X-ray wavelength.
2.4. Maintenance of cell lines
The human breast cancer cells (MDA-MB-231 and MCF-7) and RAW 264.7 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat inactivated fetal bovine serum (FBS), and 1% penicillin/streptomycin antibiotic solution and were maintained in a humidified 5% CO2 incubator at 37 °C, until the cells get adhered and achieved 80–90% confluency.2.5. Glucose estimation assay
The induction of growth inhibition and cytotoxicity in cancer cells is a well-known consequence of glucose deprivation. Considering the fact, we assessed the concentration of glucose in the culture media of MDA-MB-231 and MCF-7 cells treated with CgAgNPs (IC30, IC50 and IC70 concentrations) using Glucose estimation kit (Beacon Diagnostics, India), as per manufacturer's protocol. The absorbance of control (non-treated) and CgAgNPs treated samples were measured against blank at 505 nm. The concentration of glucose was calculated and presented in mg dl−1 using the equation below and analyzed statistically:2.6. Cell proliferation assay
Cell proliferation assay helps in investigating the quantification of viable cells and proliferation rate using crystal violet.45 In brief, the breast cancer cells, MDA-MB-231 and MCF-7 were treated with IC30, IC50 and IC70 concentrations of CgAgNPs. After 24 hours of treatment, the media was discarded and the adhered cells were fixed with absolute ethanol for 10 minutes. Further, the fixed cells were stained with 0.05% crystal violet in 20% ethanol followed by 10 minutes of incubation. Further, the cells were washed with distilled water followed by solubilization of formed crystals with methanol. The percent cell proliferation was then determined by taking absorbance at 595 nm using a microplate reader (ThermoFisher Scientific). The cell free medium was used as blank. The percent cell proliferation was calculated using the equation below:2.7. Lactate dehydrogenase (LDH) assay
LDH assay is a calorimetric cytotoxicity assay that is used to quantify the intracellular LDH released from damaged plasma membrane into the extracellular culture medium. Briefly, breast cancer cells, MDA-MB-231 and MCF-7 were treated with IC30, IC50 and IC70 concentrations of CgAgNPs. After 24 hours of treatment, the collected media was centrifuged at 3000 rpm for 3 minutes. The LDH rich supernatant was further used to estimate the LDH level using Lactate Dehydrogenase (LDH) activity assay kit (Elabscience), following the manufacturer's protocol. The absorbance of each sample was measured using microplate reader (ThermoFisher Scientific) at 450 nm. The LDH standard curve (Fig. S1†) was used to measure the level of LDH (U/L) and the obtained data was analysed statistically.2.8. Evaluation of differential gene expression by real-time quantitative PCR (qRT-PCR)
The assessment of altered expression of apoptosis related genes in CgAgNPs treated breast cancer cells, MDA-MB-231 and MCF-7 was done using qRT-PCR analysis. The breast cancer cells, MDA-MB-231 and MCF-7 were treated with IC50 concentration of CgAgNPs. The treated cells were incubated overnight and maintained in a humidified 5% CO2 atmosphere at 37 °C, followed by extraction of total RNA using TRIzol reagent. The extracted RNA was checked for its purity and concentration using Nanodrop (Thermo Scientific). The synthesis for cDNA was further done through reverse transcription of purified RNA following manufacturer's protocol provided in Thermo Scientific cDNA synthesis kit with some minor changes.46 Moreover, following MIQE standard protocol,47 qRT-PCR analysis was done with a PowerUp Thermo SYBR Green Master Mix and for the quantification, thermal cycles 5 real-time PCR system (Quant studio Thermo Fisher) was used. Different set of primers associated with apoptosis were used for screening the expression of genes (Table S1†). The mean CT values were calculated for control and CgAgNPs treated samples and analysis was done by calculating ΔΔCT value. Assessment of relative quantification of differentially expressed genes in terms of fold change was calculated by 2−ΔΔCT method.2.9. Statistical analysis
All the experiments were performed in triplicate (n = 3). The data for MTT assay, glucose estimation, cell proliferation and LDH assay were expressed as mean ± SEM. The analysis of IC30, IC50 and IC70 values for the MTT assay was done by using Graph Pad Prism 8.0.2 software. The one-way analysis of variance (one-way ANOVA) was used for the statistical analysis of all the data sets followed by Tukey's test (***p ≤ 0.001; **p ≤ 0.002; *p ≤ 0.033). The data was expressed as mean ± SEM and respective values of all groups were compared. For qRT-PCR experiment, unpaired Student's t-test (*p < 0.05, **p < 0.01, ***p < 0.001) was used and mean ± SEM of all the groups were compared to determine the statistical significance using Graph Pad Prism 8.0.2 software.3. Results and discussion
3.1. Bio-analytical techniques for the identification, characterization and evaluation of the synthesized nanoparticles
3.2. CgAgNPs exhibited potential in vitro cytotoxicity against breast cancer cells
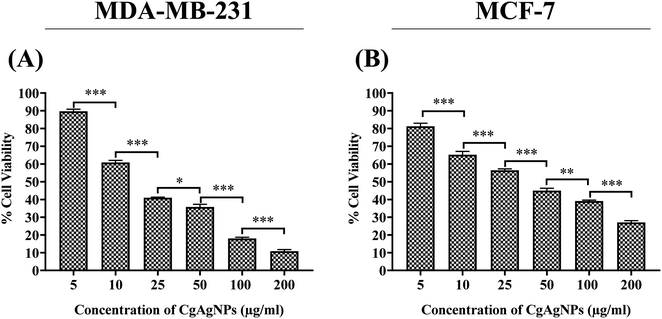
The cytotoxicity of biosynthesized silver nanoparticles was evaluated against RAW 264.7 cells and two breast cancer cells, MDA-MB-231 and MCF-7 and the inhibitory concentration at half maximal (IC50) that is required to inhibit the cancer cells proliferation by 50% as compared to the control was calculated. The anti-proliferative activity of CgAgNPs was tested at different concentrations (5, 10, 25, 50, 100 and 200 μg mL−1) using MTT assay and percentage cell viability was calculated and represented in form of histogram for RAW 264.7 cells (Fig. S4†) and breast cancer cells (Fig. 3). The result analysis clearly indicates the significant cytotoxic action of CgAgNPs against breast cancer cells, MDA-MB-231 and MCF-7 while, toxicity against RAW 264.7 cells was observed at very higher dose. The spectrophotometric analysis indicates the decrease in percent cell viability with increase in CgAgNPs concentration. For MDA-MB-231 and MCF-7 cells, the calculated IC50 value, where cell viability was reduced to 50% after CgAgNPs treatment, was found to be 18.398 ± 0.376 and 38.587 ± 1.828 μg mL−1, respectively, however, for RAW 264.7 cells it was found to be 195.221 ± 2.394 μg mL−1. The synthesized CgAgNPs showed high cytotoxicity to MDA-MB-231 and MCF-7 cells at low dose while represent biosafety towards healthy cells. Moreover, IC30 and IC70 values of CgAgNPs were also calculated as 8.487 ± 0.065 and 65.678 ± 2.571 μg mL−1, respectively for MDA-MB-231 cells and for MCF-7 it was reported as 8.839 ± 0.398 (IC30) and 180.61 ± 4.551 μg mL−1 (IC70). Various mechanisms contribute to the cytotoxic activity of the synthesized CgAgNPs, including cellular disintegration, buildup of free radicals (ROS & RNS), damage to nucleic acid and degradation of surface protein. As the wrapped silver particle (Ag) get released from the bounded fungal filtrate moiety, they start piling up on the targeted sites that obstructs in electron transport system ultimately causing efflux of intracellular ion and damage to cell.76 A previous study has reported in accordance with the present data that depicts the potential cytotoxic action at IC50 value (8.7 μg mL−1) against MDA MB-231 cells treated with biosynthesized AgNPs from B. funiculus.77 The synthesis of AgNPs from leaf extract of C. edulis exhibited concentration dependent cytotoxicity on mammalian cells (L929).78 Considering the cytotoxic potential of CgAgNPs against breast cancer cells, subsequent experiments have been performed to study the anti-proliferative potential of CgAgNPs.3.3. CgAgNPs inhibited glucose uptake in breast cancer cells
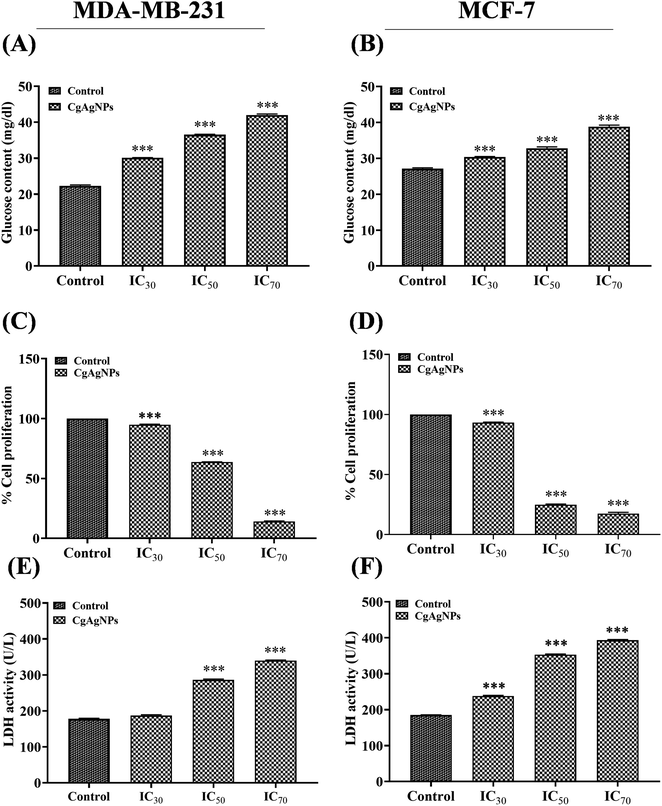
The quantification of glucose content in culture media of CgAgNPs treated breast cancer cells was done to evaluate the metabolic alteration induced by CgAgNPs. After treatment with the respective concentration of CgAgNPs (IC30, IC50, and IC70) compared with respect to control, the glucose content was significantly increased in a dose dependent manner (Fig. 4A and B). As glucose deprivation adversely inhibits the cell growth, therefore, the amount of glucose present in culture media of CgAgNPs treated breast cancer cells was analyzed that helps in the quantification of the glucose consumption. The culture media of control groups for both the cells, MDA-MB-231 and MCF-7 exhibited very low glucose content, measuring 22.309 ± 0.238 and 27.117 ± 0.235 mg dl−1, respectively as compared to treated group. The glucose content in culture media of MDA-MB-231, administered with different doses of CgAgNPs (IC30, IC50 and IC70) were found to be 30.096 ± 0.063, 36.539 ± 0.146 and 42.021 ± 0.296 mg dl−1, respectively, whereas in culture media of MCF-7 cells was found to be 30.384 ± 0.122, 32.789 ± 0.409 and 38.846 ± 0.409 mg dl−1, respectively. The result analysis revealed that in culture media of breast cancer cells treated with CgAgNPs at higher dose of IC70 exhibits high glucose content, indicating reduced cell viability, proliferation and altered metabolic activity. Previous findings have reported that AgNPs substantially altered the glucose metabolism in hepatoma cells (HepG2) by reducing the glucose consumption cancer cells.79 The glucose availability in AgNPs treated HepG2 cells has shown that low level of glucose supply induces ROS generation in cancer cells.80 Hence, the finding suggest that reduced glucose uptake may act as regulatory point to precisely target tumor cells while reducing adverse effects on healthy cells.3.4. CgAgNPs significantly inhibited cell proliferation in breast cancer cells
The cell proliferation assay by crystal violet dye that generally stains adherent cells helps in the determination of proliferation and viability of the cultured cells. Upon treatment with the different concentration of CgAgNPs, the adhered cells undergo cell death and lose adherence. The concentration-dependent cell abundance was determined by crystal violet dye that generally binds to nucleic acid and proteins of adhered cultured cells. This leads to indirect quantification of non-viable cells and helps in determination of cell proliferation rate. Cells that have undergone cell death, lose adherence property and therefore significantly colour intensity of crystal violet dye is reduced.81 According to the fact, after treatment of MDA-MB-231 and MCF-7 cells with respective IC30, IC50, and IC70 concentration of CgAgNPs, we further sighted to determine the percent cell proliferation of the adhered cells. The CgAgNPs at different inhibitory concentration of IC30, IC50 and IC70 showed dose-dependent decrease in cell proliferation rate in breast cancer cells (Fig. 4C and D) which was in accordance with the result of MTT assay. For MDA-MB-231 cells, the percent cell proliferation treated with IC30, IC50 and IC70 concentrations of CgAgNPs was found to be 94.878 ± 0.459, 63.63 ± 0.230 and 13.98 ± 0.483%, respectively as compared to control. Whereas, for MCF-7 cells, the percent cell proliferation was found to be 93.183 ± 0.426, 24.708 ± 0.661 and 17.463 ± 0.571%, respectively. Cell proliferation is a highly regulated process of normal cells. Lack of contact inhibition, alters protein expression related to cell cycle as well as morphological transitions of epithelial cells contribute to uncontrolled proliferation of cancer cells.82 Similarly, previous researches has shown HCT116 cells treated with AgNPs exhibited significant anti-proliferative action.83 The AgNPs synthesized from Abutilon indicum exhibited remarkable anti-proliferative action against colon cancer cells (COLO 205).84 Collectively, the findings revealed that CgAgNPs induces significant decrease in cell proliferation in a concentration dependent manner.3.5. CgAgNPs enhanced LDH release in breast cancer cells
LDH assay determines the cytotoxic effect against the breast cancer cells and degradation of cellular integrity after treatment with CgAgNPs. The effect of CgAgNPs treatment on LDH activity in culture media of breast cancer cells was monitored by changes in absorbance at 450 nm during NAD+ reduction. Compared to control, the treated groups showed more toxicity as validated by high LDH activity in culture media (Fig. 4E and F). CgAgNPs treated breast cancer cells (MDA-MB-231 and MCF-7) causes decreased cytosolic LDH level which shows decrease in proliferation rate in a dose dependent manner. CgAgNPs induces damage to the membrane of cancer cell that leads to the release of cytosolic LDH to the supernatant. The treated cells displayed high level of released LDH as compared to the untreated cells. CgAgNPs at different concentration of IC30, IC50 and IC70 showed dose-dependent rise of LDH activity in culture media of breast cancer cells, MDA-MB-231 and MCF-7. In the culture media of MDA-MB-231 cells, the LDH activity at different concentrations of CgAgNPs that is IC30, IC50 and IC70 was found to be 186.731 ± 1.411, 286.084 ± 2.334 and 339.806 ± 1.483 U L−1, respectively as compared to control (177.67 ± 1.682 U L−1). Whereas, for culture media of CgAgNPs treated MCF-7 cells, the LDH activity at different concentrations of IC30, IC50 and IC70 was found to be 237.864 ± 1.682, 353.074 ± 1.802 and 393.204 ± 2.201 U L−1, respectively as compared to control (184.79 ± 0.856 U L−1). During cellular damage, LDH being a soluble cytosolic enzyme gets released into the media and that is indicative of increased cytotoxicity. LDH promotes the conversion of lactate into pyruvate via reduction of NAD+ into NADH which further reacts with chromogenic substrate that show colour change. The histogram obtained for LDH activity and MTT assay reveals inverse relationship that validates the cytotoxicity of bio-fabricated CgAgNPs. In both MTT and LDH assay, with rise in CgAgNPs concentration we found substantial rise in cytotoxic level which was confirmed by measuring percent cell viability and LDH level, respectively. Previous studies has revealed the potential cytotoxic action of AgNPs on lung cancer cells (A549) that caused damage to cell membrane with LDH release in a dose dependent manner.85 The biogenically synthesized AgNPs from L. boronitolerans has shown significant cytotoxic action against C2C12 muscle cell line that leads to distortion of cellular membrane in a concentration dependent manner.86 Therefore, the result validates that green synthesized nanoparticles CgAgNPs can serve as a potential cytotoxic as well as anti-proliferative agent against the breast cancer cells.3.6. CgAgNPs treated breast cancer cells exhibited altered gene expression
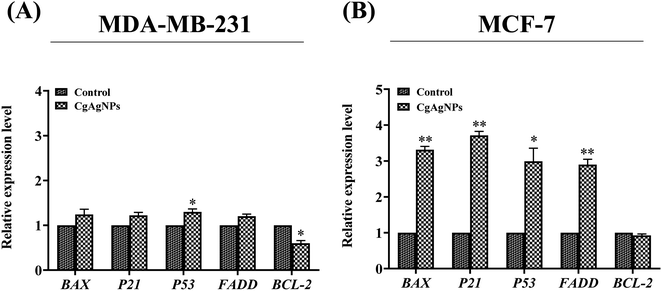
Apoptosis is a key regulator of tissue homeostasis as well as highly regulates the process of tumorigenesis by causing cell death.87 The breast cancer cells (MDA-MB-231 and MCF-7) treated with CgAgNPs were investigated for the differentially expressed genes in control and treated groups. The molecular approach guiding CgAgNPs induced apoptosis in MDA-MB-231 and MCF-7 cells were determined by examining the fold change expression levels of apoptosis related genes (BAX, BCL-2, P21, P53 and FADD) as shown in Fig. 5A and B. The qRT-PCR data analysis has revealed upregulation of pro-apoptotic gene, BAX in MDA-MB-231 and MCF-7 cells with fold increase of 1.242 ± 0.117 (p-value 0.175) and 3.315 ± 0.093 (p-value 0.002), respectively with respect to control. In MCF-7 cells, the BAX gene expression shows statistically significant difference between control group and treated group. The cell cycle growth regulator, P21 was found to be upregulated with fold increase of 1.223 ± 0.065 (p-value 0.075) and 3.714 ± 0.112 (p-value 0.002) in MDA-MB-231 and MCF-7 cells, respectively with respect to control, showing statistically significant difference in MCF-7 cells only. The tumor suppressor gene, P53 showed upregulation with fold increase of 1.299 ± 0.065 (p-value 0.044) and 2.994 ± 0.363 (p-value 0.032) in MDA-MB-231 and MCF-7 cells, respectively showing statistical difference in both the treated cells with respect to control. In MDA-MB-231 and MCF-7 cells, the FADD gene also gets upregulated with respect to control and having fold increase of 1.202 ± 0.049 (p-value 0.054) and 2.899 ± 0.149 (p-value 0.006), respectively showing statistically significant difference in MCF-7 cells only. CgAgNPs treated cells showed enhanced expression of P53 that possibly indicates activation of apoptosis signaling pathway through conformational changes in amino terminal of mitochondrial membrane that triggers release of cytochrome c.88 The anti-apoptotic gene, BCL-2 shows downregulation by 0.599 ± 0.061 (p-value 0.022) and 0.924 ± 0.041 (p-value 0.205) fold in MDA-MB-231 and MCF-7 cells, respectively showing statistically significant difference in MDA-MB-231 cells only with respect to control. The tumor suppressor gene, P53 modulate the process of apoptosis by its activation through regulation of Bcl-2 family members. These protein family comprises both pro-apoptotic gene, BAX as well as anti-apoptotic gene BCL-2.89 The heterodimerization of Bax (Bcl-2 associated X) protein and Bcl-2 shows structural sequence homology that triggers apoptosis by causing membrane permeabilization in mitochondria causing release of cytochrome c into the cytosol that further stimulate caspases and p53 to induce apoptosis.90 The downregulation of BCL-2 by CgAgNPs suggests cell death by directly binding and suppressing pro-apoptotic Bcl-2 family proteins through mitochondrial dependent intrinsic cascade pathway. CgAgNPs could be possibly involved in the upregulation of BAX gene expression that leads to its activation and dimerization at the outer membrane of mitochondria causing permeabilization and ultimately leading to apoptosis. Bcl-2 act as an antagonist in the apoptotic machinery pathway whereas Bax promotes apoptosis acting as agonists by competing with Bcl-2.91 Thus, in response to nuclear damage, the tumor suppressor protein p53 targets both Bcl-2 and Bax which triggers apoptosis and cell cycle inhibition.92 Hence, CgAgNPs treated breast cancer cells may prove to be supporting evidence for inducing apoptotic cell death and provide an efficient way in combating progression of cancer. The p21, oncogenic cell cycle inhibitor protein mediates growth inhibition via p53 dependent apoptotic pathway. p21 inhibit CDKs (cyclin dependent kinases) which are essential for cell cycle growth.93 Thus, its upregulation promotes regression of breast cancer cells. Apoptosis or PCD is also regulated by Fas mediated signaling. The acquisition of FADD triggers Fas mediated apoptosis which recruits and activate caspase-8 to form homodimers and transmit death signal through caspase-3.94 The cleavage of BH3-interacting domain death agonist (Bid) by caspase-8 causes porosity in mitochondrial membrane leading to cytosolic release of cytochrome c, thus ultimately leads to activation of downstream caspases on interaction with apoptotic protease activating factor (Apaf).95 Thus, the present result demonstrates that the CgAgNPs treated breast cancer cells, MDA-MB-231 and MCF-7 cells triggers differential expression of proapoptotic genes (BAX, P21 and P53), antiapoptotic gene (BCL-2) and FADD gene responsible for intrinsic as well as extrinsic apoptotic pathway. Previous study has evidenced that the green mediated synthesis of AgNPs from Gossypium hirsutum induces apoptosis in A549 lung cancer cells through down regulation of BCL-2 and enhanced expression of BAX genes.88 The AgNPs mediated synthesis from aqueous extract of Bergenia ligulata in MCF-7 leads to cell cycle arrest due to upregulation of phosphorylated p53 and ROS generation that ultimately induces apoptosis.96 Thus, the present study has been found in accordance with the previous researches and shows the potential chemotherapeutic action of CgAgNPs and role of apoptosis mediated biomarker genes on treated breast cancer cells.4. Conclusion
Fungal endophyte mediated green silver nanoparticles have provided striking approach facilitating for biomedical and therapeutic interventions. The present study revealed the fungal endophyte C. gloeosporioides isolated from the O. indicum as a potential source for the fabrication of green silver nanoparticles exhibiting potential cytotoxic and anti-proliferative activities against human breast cancer cells. The preliminary detection showed a visible colour change and UV- Visible spectral analysis showed characteristic peak in the wavelength range of 400–450 nm. The FT-IR spectroscopy analysis depicts the presence of functionalized fungal biomolecules carrying hydroxyl, carbonyl and free amino group that might be responsible for capping and stabilizing the silver nanoparticles. The crystallite size and purity were further confirmed by XRD analysis. The microscopic techniques such as AFM, FESEM and TEM depicts the agglomeration, size, surface topology as well as shape of the synthesized CgAgNPs. Further, CgAgNPs exhibited significant antiproliferative activity against human breast cancer cells, MDA-MB-231 and MCF-7 that was observed through reduction in cell viability in a dose-dependent manner. The cytotoxic activity of CgAgNPs against breast cancer cells was further validated through alteration in biochemical parameters. The reduced glucose uptake, reduced cell proliferation, and increased release of LDH in CgAgNPs treated breast cancer cells further confirms its cytotoxic and antiproliferative activity. The gene expression analysis revealed the upregulation of pro-apoptotic genes (BAX and FADD) cell cycle inhibitor gene (P21) and tumor suppressor gene (P53), while downregulation of anti-apoptotic gene (BCL-2). Collectively, this differential expression of genes provides the compelling evidence for the apoptosis inducing potential of the synthesized nanoparticle CgAgNPs. In summary, the synthesized CgAgNPs exhibit promising antiproliferative and cytotoxic effects against human breast cancer cells, highlighting their significant potential for translational studies in animal models and for the development of nano-based drugs.Author contributions
Priyamvada Gupta: data curation; formal analysis; investigation; methodology; roles/writing-original draft; writing-review & editing; visualization. Swati Singh: data curation; formal analysis; investigation; methodology; roles/writing-original draft; writing review & editing; visualization. Nilesh Rai: data curation; formal analysis; investigation; writing review & editing. Ashish Verma: data curation; formal analysis; investigation; writing review & editing. Harshita Tiwari: data curation; formal analysis; investigation; writing review & editing. Swapnil C. Kamble: Formal analysis; Data curation. Hemant Kumar Gautam: formal analysis; writing review and editing. Vibhav Gautam: conceptualization; methodology; formal analysis; funding acquisition; investigation; project administration; resources; software; writing review and editing.Conflicts of interest
The corresponding author on behalf of all the authors declares no potential conflict of interest.Acknowledgements
PG would like to thank Science and Engineering Research Board, India (EMEQ scheme; EEQ/2019/000025) and Banaras Hindu University, Varanasi, India for financial support. SS and NR are thankful to University Grants Commission, New Delhi, India for the financial support. AV is thankful to Council of Scientific and Industrial Research, New Delhi, India for the Junior Research Fellowship. The corresponding author, on behalf of all the authors, acknowledges Central Instrumental Facility (CIF) at IIT (BHU) in Varanasi, India, Department of Physics (BHU) as well as the Sophisticated Central Instrumentation Facility (SCIF) at IIT Dharwad, India. VG would like to acknowledge Dr Rajiv Kumar from Centre of Experimental Medicine and Surgery, IMS (BHU), Varanasi, for providing RAW 264.7 cells. VG expresses gratitude to Dr Shyam Lal M from INST Mohali, India, for his valuable contributions throughout the revision process. The VG laboratory receives funding from the Science and Engineering Research Board (SERB)-EMEQ grant (EEQ/2019/000025), the University Grants Commission in New Delhi, India, and internal funding from Banaras Hindu University in Varanasi, India, under the Institution of Eminence Scheme (Seed and Bridge Grant). The figure of graphical abstract was created with help of http://www.biorender.com/.References
- S. Bayda, M. Adeel, T. Tuccinardi, M. Cordani and F. Rizzolio, Molecules, 2019, 25, 112 CrossRef PubMed.
- H. Barabadi, M. A. Mahjoub, B. Tajani, A. Ahmadi, Y. Junejo and M. Saravanan, J. Cluster Sci., 2019, 30, 259–279 CrossRef CAS.
- H. Tiwari, N. Rai, S. Singh, P. Gupta, A. Verma, A. K. Singh, Kajal, P. Salvi, S. K. Singh and V. Gautam, Bioengineering, 2023, 10, 760 CrossRef CAS PubMed.
- S. Ahmadi, M. Fazilati, S. M. Mousavi and H. Nazem, J. Exp. Nanosci., 2020, 15, 363–380 CrossRef CAS.
- P. Gupta, N. Rai, A. Verma and V. Gautam, Med. Res. Rev., 2023, 44, 138–168 CrossRef PubMed.
- A. Malhotra, K. Dolma, N. Kaur, Y. S. Rathore, Ashish, S. Mayilraj and A. R. Choudhury, Bioresour. Technol., 2013, 142, 727–731 CrossRef CAS PubMed.
- N. Rai, P. Kumari Keshri, A. Verma, S. C. Kamble, P. Mishra, S. Barik, S. Kumar Singh and V. Gautam, Mycology, 2021, 12, 139–159 CrossRef CAS PubMed.
- J. Wen, S. K. Okyere, S. Wang, J. Wang, L. Xie, Y. Ran and Y. Hu, J. Fungi, 2022, 8, 205 CrossRef CAS PubMed.
- P. K. Keshri, N. Rai, A. Verma, S. C. Kamble, S. Barik, P. Mishra, S. K. Singh, P. Salvi and V. Gautam, Mycol. Prog., 2021, 20, 577–594 CrossRef.
- N. Atri, N. Rai, A. K. Singh, M. Verma, S. Barik, V. Gautam and S. K. Singh, J. Sci. Res., 2020, 64, 127–133 Search PubMed.
- A. Verma, N. Rai, S. C. Kamble, P. Mishra, S. Barik, R. Kumar, S. K. Singh, P. Salvi and V. Gautam, Traditional Medicine for Neuronal Health, 2023, 82–102 Search PubMed.
- R. Kousar, M. Naeem, M. I. Jamaludin, A. Arshad, A. N. Shamsuri, N. Ansari, S. Akhtar, A. Hazafa, J. Uddin, A. Khan and A. Al-Harrasi, Am. J. Cancer Res., 2022, 12, 2897–2919 CAS.
- A. Verma, N. Rai, P. Gupta, S. Singh, H. Tiwari, S. B. Chauhan, V. Kailashiya and V. Gautam, Environ. Toxicol., 2023, 38, 2509–2523 CrossRef CAS PubMed.
- N. Rai, P. Gupta, P. K. Keshri, A. Verma, P. Mishra, D. Kumar, A. Kumar, S. K. Singh and V. Gautam, Appl. Biochem. Biotechnol., 2022, 194, 3296–3319 CrossRef CAS PubMed.
- A. Verma, P. Gupta, N. Rai, R. K. Tiwari, A. Kumar, P. Salvi, S. C. Kamble, S. K. Singh and V. Gautam, J. Fungi, 2022, 8, 285 CrossRef CAS PubMed.
- P. Gupta, A. Verma, N. Rai, A. K. Singh, S. K. Singh, B. Kumar, R. Kumar and V. Gautam, ACS Chem. Biol., 2021, 16, 2068–2086 CrossRef CAS PubMed.
- S. Barik, N. Rai, P. Mishra, S. K. Singh and V. Gautam, Curr. Sci., 2020, 118, 698–699 Search PubMed.
- N. Rai, P. Gupta, A. Verma, S. Singh, H. Tiwari, R. Kumar, S. K. Singh and V. Gautam, in Handbook of Oncobiology: From Basic to Clinical Sciences, ed. R. C. Sobti, N. K. Ganguly and R. Kumar, Springer Nature Singapore, Singapore, 2023, pp. 1–24, DOI:10.1007/978-981-99-2196-6_70-1.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 CrossRef CAS PubMed.
- A. Gade, P. Bonde, A. Ingle, P. Marcato, N. Duran and M. Rai, J. Biobased Mater. Bioenergy, 2008, 2, 243–247 CrossRef.
- V. Ahluwalia, J. Kumar, R. Sisodia, N. A. Shakil and S. Walia, Ind. Crops Prod., 2014, 55, 202–206 CrossRef CAS.
- T. Akther, V. Mathipi, N. S. Kumar, M. Davoodbasha and H. Srinivasan, Environ. Sci. Pollut. Res., 2019, 26, 13649–13657 CrossRef CAS PubMed.
- M. Govindappa, M. Lavanya, P. Aishwarya, K. Pai, P. Lunked, B. Hemashekhar, B. Arpitha, Y. Ramachandra and V. B. Raghavendra, Bionanosci., 2020, 10, 928–941 CrossRef.
- U. Munawer, V. B. Raghavendra, S. Ningaraju, K. L. Krishna, A. R. Ghosh, G. Melappa and A. Pugazhendhi, Int. J. Pharm., 2020, 588, 119729 CrossRef CAS PubMed.
- N. A. Razak, N. Abu, W. Y. Ho, N. R. Zamberi, S. W. Tan, N. B. Alitheen, K. Long and S. K. Yeap, Sci. Rep., 2019, 9, 1514 CrossRef PubMed.
- K. Nohara, F. Wang and S. Spiegel, Breast Cancer Res. Treat., 1998, 48, 149–157 CrossRef CAS PubMed.
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, CA Cancer J. Clin., 2022, 72, 7–33 CrossRef PubMed.
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, CA Cancer J. Clin., 2021, 71, 7–33 CrossRef PubMed.
- P. S. Ward and C. B. Thompson, Cancer cell, 2012, 21, 297–308 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- M. A. Khajah, S. Khushaish and Y. A. Luqmani, PLoS One, 2022, 17, e0272449 CrossRef CAS PubMed.
- R. L. Aft, F. W. Zhang and D. Gius, Br. J. Cancer, 2002, 87, 805–812 CrossRef CAS PubMed.
- Y. Feng, Y. Xiong, T. Qiao, X. Li, L. Jia and Y. Han, Cancer Med., 2018, 7, 6124–6136 CrossRef PubMed.
- P. Miao, S. Sheng, X. Sun, J. Liu and G. Huang, IUBMB life, 2013, 65, 904–910 CrossRef CAS PubMed.
- S. Kim, J. E. Choi, J. Choi, K.-H. Chung, K. Park, J. Yi and D.-Y. Ryu, Toxicol. In Vitro, 2009, 23, 1076–1084 CrossRef CAS PubMed.
- Y. G. Yuan, S. Zhang, J. Y. Hwang and I. K. Kong, Oxid. Med. Cell. Longevity, 2018, 2018, 6121328 Search PubMed.
- K. L. Thu, I. Soria-Bretones, T. W. Mak and D. W. Cescon, Cell cycle (Georgetown, Tex.), 2018, 17, 1871–1885 CrossRef CAS PubMed.
- D. Acharya, S. Satapathy, J. J. Thathapudi, P. Somu and G. Mishra, Mater. Technol., 2022, 37, 569–580 CrossRef CAS.
- S. Zhang, H. Jiang, B. Gao, W. Yang and G. Wang, Front. Cell Dev. Biol., 2021, 9, 811585 CrossRef PubMed.
- S. Asad, S. Supriya, C. Gopal and A. Absar, Spectrochim. Acta, Part A, 2013, 114, 144–147 CrossRef PubMed.
- H. Bagur, R. S. Medidi, P. Somu, P. J. Choudhury, C. S. Karua, P. K. Guttula, G. Melappa and C. C. Poojari, Mater. Technol., 2022, 37, 167–178 CrossRef CAS.
- R. S. Yehia and H. Al-Sheikh, World J. Microbiol. Biotechnol., 2014, 30, 2797–2803 CrossRef CAS PubMed.
- N. Rai, P. K. Keshri, P. Gupta, A. Verma, S. C. Kamble, S. K. Singh and V. Gautam, PLoS One, 2022, 17, e0264673 CrossRef CAS PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- D. P. Bonfim, C. V. Nakamura, J. X. de Araújo Júnior, G. L. Pessini, P. E. C. Leite, J. A. Morgado-Díaz and F. J. P. R. Leve, Phytother. Res., 2021, 35, 3769–3780 CrossRef CAS PubMed.
- V. Gautam, A. Singh, S. Yadav, S. Singh, P. Kumar, S. Sarkar Das and A. K. J. D. Sarkar, Development, 2021, 148, dev190033 CAS.
- S. A. Bustin, V. Benes, J. A. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, M. W. Pfaffl, G. L. Shipley, J. Vandesompele and C. T. Wittwer, Clin. Chem., 2009, 55, 611–622 CrossRef CAS PubMed.
- P. Mulvaney, Langmuir, 1996, 12, 788–800 CrossRef CAS.
- C. Krishnaraj, E. G. Jagan, S. Rajasekar, P. Selvakumar, P. T. Kalaichelvan and N. Mohan, Colloids Surf., B, 2010, 76, 50–56 CrossRef CAS PubMed.
- N. Rai, P. Gupta, A. Verma, S. K. Singh and V. Gautam, BioFactors, 2023, 49, 663–683 CrossRef CAS PubMed.
- N. Rai, P. Gupta, A. Verma, R. K. Tiwari, P. Madhukar, S. C. Kamble, A. Kumar, R. Kumar, S. K. Singh and V. Gautam, ACS omega, 2023, 8, 3768–3784 CrossRef CAS PubMed.
- S. Anil Kumar, M. K. Abyaneh, S. W. Gosavi, S. K. Kulkarni, R. Pasricha, A. Ahmad and M. I. Khan, Biotechnol. Lett., 2007, 29, 439–445 CrossRef CAS PubMed.
- H. Korbekandi, Z. Ashari, S. Iravani and S. Abbasi, Iran. J. Pharm. Res., 2013, 12, 289–298 CAS.
- A. Ingle, A. Gade, S. Pierrat, C. Sonnichsen and M. Rai, Curr. Nanosci., 2008, 4, 141–144 CrossRef CAS.
- N. Vigneshwaran, N. M. Ashtaputre, P. V. Varadarajan, R. P. Nachane, K. M. Paralikar and R. H. Balasubramanya, Mater. Lett., 2007, 61, 1413–1418 CrossRef CAS.
- K. Anandalakshmi, J. Venugobal and V. Ramasamy, Appl. Nanosci., 2016, 6, 399–408 CrossRef CAS.
- R. Anith Jose, D. Devina Merin, T. S. Arulananth and N. Shaik, J. Nanomater., 2022, 2022, 4056551 Search PubMed.
- P. Khandel and S. K. Shahi, J. Nanostruct. Chem., 2018, 8, 369–391 CrossRef CAS.
- P. Azmath, S. Baker, D. Rakshith and S. Satish, Saudi Pharm. J., 2016, 24, 140–146 CrossRef PubMed.
- K. Prabakaran, C. Ragavendran and D. Natarajan, RSC Adv., 2016, 6, 44972–44986 RSC.
- K. I. Alsamhary, Saudi J. Biol. Sci., 2020, 27, 2185–2191 CrossRef CAS PubMed.
- A. Mahajan, A. Arya and T. S. Chundawat, Synth. Commun., 2019, 49, 1926–1937 CrossRef CAS.
- D. Arumai Selvan, D. Mahendiran, R. Senthil Kumar and A. Kalilur Rahiman, J. Photochem. Photobiol., B, 2018, 180, 243–252 CrossRef CAS PubMed.
- J. Coates, Encyclopedia of Analytical Chemistry, 2000, 10815–10837 Search PubMed.
- D. Singh, V. Rathod, S. Ninganagouda, J. Hiremath, A. K. Singh and J. Mathew, Bioinorg. Chem. Appl., 2014, 2014, 408021 Search PubMed.
- A. Gole, C. Dash, V. Ramakrishnan, S. Sainkar, A. Mandale, M. Rao and M. Sastry, Langmuir, 2001, 17, 1674–1679 CrossRef CAS.
- D. Wang, B. Xue, L. Wang, Y. Zhang, L. Liu and Y. Zhou, Sci. Rep., 2021, 11, 10356 CrossRef CAS PubMed.
- M. A. Yassin, A. M. Elgorban, A. E.-R. M. A. El-Samawaty and B. M. A. Almunqedhi, Saudi J. Biol. Sci., 2021, 28, 2123–2127 CrossRef CAS PubMed.
- V. S. Kotakadi, S. A. Gaddam, S. K. Venkata, P. V. G. K. Sarma and D. V. R. Sai Gopal, 3 Biotech, 2016, 6, 216 CrossRef PubMed.
- T. Singh, K. Jyoti, A. Patnaik, A. Singh, R. Chauhan and S. S. Chandel, J. Genet. Eng. Biotechnol., 2017, 15, 31–39 CrossRef PubMed.
- G. Li, D. He, Y. Qian, B. Guan, S. Gao, Y. Cui, K. Yokoyama and L. Wang, Int. J. Mol. Sci., 2012, 13, 466–476 CrossRef CAS PubMed.
- P. Gupta, N. Rai, A. Verma, D. Saikia, S. P. Singh, R. Kumar, S. K. Singh, D. Kumar and V. Gautam, ACS omega, 2022, 7, 46653–46673 CrossRef CAS PubMed.
- N. Ahmad, S. Sharma, M. K. Alam, V. N. Singh, S. F. Shamsi, B. R. Mehta and A. Fatma, Colloids Surf., B, 2010, 81, 81–86 CrossRef CAS PubMed.
- M. Manjunath Hulikere and C. G. Joshi, Process Biochem., 2019, 82, 199–204 CrossRef CAS.
- P. Salvi, H. Mahawar, R. Agarrwal, V. Gautam and R. Deshmukh, Front. Microbiol., 2022, 13, 3407 Search PubMed.
- H. Du, T. M. Lo, J. Sitompul and M. W. Chang, Biochem. Biophys. Res. Commun., 2012, 424, 657–662 CrossRef CAS PubMed.
- S. Gurunathan, J. W. Han, V. Eppakayala, M. Jeyaraj and J.-H. J. B. R. I. Kim, BioMed Res. Int., 2013, 2013, 1–10 CrossRef PubMed.
- S. V. Otari, S. H. Pawar, K. S. P. Sanjay, K. S. Raushan, K. Sang-Yong, L. Jai Hyo and Z. Liaoyuan, J. Microbiol. Biotechnol., 2017, 27, 731–738 CrossRef CAS PubMed.
- M. J. Lee, S. J. Lee, S. J. Yun, J.-Y. Jang, H. Kang, K. Kim, I.-H. Choi and S. Park, Int. J. Nanomed., 2016, 55–68 CAS.
- M. Zuberek, D. Wojciechowska, D. Krzyzanowski, S. Meczynska-Wielgosz, M. Kruszewski and A. Grzelak, J. Nanobiotechnol., 2015, 13, 72 CrossRef PubMed.
- M. Feoktistova, P. Geserick and M. Leverkus, Cold Spring Harb. Protoc., 2016, 2016, 343–346 Search PubMed.
- M. A. Feitelson, A. Arzumanyan, R. J. Kulathinal, S. W. Blain, R. F. Holcombe, J. Mahajna, M. Marino, M. L. Martinez-Chantar, R. Nawroth, I. Sanchez-Garcia, D. Sharma, N. K. Saxena, N. Singh, P. J. Vlachostergios, S. Guo, K. Honoki, H. Fujii, A. G. Georgakilas, A. Bilsland, A. Amedei, E. Niccolai, A. Amin, S. S. Ashraf, C. S. Boosani, G. Guha, M. R. Ciriolo, K. Aquilano, S. Chen, S. I. Mohammed, A. S. Azmi, D. Bhakta, D. Halicka, W. N. Keith and S. Nowsheen, Semin. Cancer Biol., 2015, 35(Suppl), S25–s54 CrossRef PubMed.
- S. Gurunathan, M. Qasim, C. Park, H. Yoo, J.-H. Kim and K. Hong, BioMed Res. Int., 2018, 19, 2269 Search PubMed.
- R. Mata, J. R. Nakkala and S. R. Sadras, Colloids Surf., B, 2015, 128, 276–286 CrossRef CAS PubMed.
- Y. S. Lee, D. W. Kim, Y. H. Lee, J. H. Oh, S. Yoon, M. S. Choi, S. K. Lee, J. W. Kim, K. Lee and C.-W. Song, Arch. Toxicol., 2011, 85, 1529–1540 CrossRef CAS PubMed.
- D. Bhatia, A. Mittal and D. K. Malik, Indian J. Exp. Biol., 2021, 15, 427–440 Search PubMed.
- J. Tower, Ageing Res. Rev., 2015, 23, 90–100 CrossRef CAS PubMed.
- N. Kanipandian, D. Li and S. Kannan, Biotechnol. Rep., 2019, 23, e00339 CrossRef PubMed.
- M. A. Anderson, J. Deng, J. F. Seymour, C. Tam, S. Y. Kim, J. Fein, L. Yu, J. R. Brown, D. Westerman and E. G. Si, Blood, The Journal of the American Society of Hematology, 2016, 127, 3215–3224 CAS.
- F. Bray, J. S. Ren, E. Masuyer and J. Ferlay, Int. J. Cancer, 2013, 132, 1133–1145 CrossRef CAS PubMed.
- J. M. Hardwick and L. Soane, Cold Spring Harbor Perspect. Biol., 2013, 5, a008722 Search PubMed.
- A. Basu and S. Haldar, Mol. Hum. Reprod., 1998, 4, 1099–1109 CrossRef CAS PubMed.
- J. W. Harper, G. R. Adami, N. Wei, K. Keyomarsi and S. Elledge, Cell, 1993, 75, 805–816 CrossRef CAS PubMed.
- A. M. Chinnaiyan, K. O'Rourke, M. Tewari and V. M. Dixit, Cell, 1995, 81, 505–512 CrossRef CAS PubMed.
- K. Huang, J. Zhang, K. L. O'Neill, C. B. Gurumurthy, R. M. Quadros, Y. Tu and X. Luo, J. Biol. Chem., 2016, 291, 11843–11851 CrossRef CAS PubMed.
- P. Takáč, R. Michalková, M. Čižmáriková, Z. Bedlovičová, Ľ. Balážová and G. Takáčová, Life, 2023, 13 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. Table S1. List of forward and reverse primers, and their sequences used in qRT-PCR analysis. Table S2. FTIR analysis of CgAgNPs. Fig. S1. Calibration curve of pyruvic acid for LDH assay. Fig. S2. FTIR spectrum of the aqueous extract of C. gloeosporioides. Fig. S3. Two dimensional (2D) AFM image showing the surface topography and dispersity of CgAgNPs. Fig. S4: Effect of CgAgNPs on viability of RAW 264.7 cells determined through MTT assay. The data is representative of three independent experiments. The statistical significance was determined through calculation of p-value where mean ± SEM of percentage cell viability of RAW 264.7 cells were compared using one-way ANOVA followed by Tukey. The statistical significance is represented as: ***p ≤ 0.001; **p ≤ 0.002; and *p ≤ 0.033. See DOI: https://doi.org/10.1039/d3ra06145k |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2024 |