Open Access Article
Open Access ArticleDroplet microfluidic system for high throughput and passive selection of bacteria producing biosurfactants†
Klaudia
Staskiewicz
 ac,
Maria
Dabrowska-Zawada
ac,
Maria
Dabrowska-Zawada
 a,
Lukasz
Kozon
a,
Lukasz
Kozon
 bd,
Zofia
Olszewska
bd,
Zofia
Olszewska
 b,
Lukasz
Drewniak
b,
Lukasz
Drewniak
 a and
Tomasz S.
Kaminski
a and
Tomasz S.
Kaminski
 *ab
*ab
aDepartment of Environmental Microbiology and Biotechnology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Poland. E-mail: ts.kaminski2@uw.edu.pl
bDepartment of Molecular Biology, Institute of Biochemistry, Faculty of Biology, University of Warsaw, Poland
cDioscuri Centre for Physics and Chemistry of Bacteria, Institute of Physical Chemistry, Polish Academy of Sciences, Warsaw, Poland
dInstitute of Physical Chemistry, Polish Academy of Sciences, Warsaw, Poland
First published on 4th March 2024
Abstract
Traditional methods for the enrichment of microorganisms rely on growth in a selective liquid medium or on an agar plate, followed by tedious characterization. Droplet microfluidic techniques have been recently used to cultivate microorganisms and preserve enriched bacterial taxonomic diversity. However, new methods are needed to select droplets comprising not only growing microorganisms but also those exhibiting specific properties, such as the production of value-added compounds. We describe here a droplet microfluidic screening technique for the functional selection of biosurfactant-producing microorganisms, which are of great interest in the bioremediation and biotechnology industries. Single bacterial cells are first encapsulated into picoliter droplets for clonal cultivation and then passively sorted at high throughput based on changes in interfacial tension in individual droplets. Our method expands droplet-based microbial enrichment with a novel approach that reduces the time and resources needed for the selection of surfactant-producing bacteria.
Introduction
Microorganisms produce metabolites that exhibit a variety of biological activities. Many of them are hormones, or compounds with antimicrobial, antitumor, herbicidal, pigment, and insecticidal properties. There has been increasing research and industrial interest in bacterial surfactants – amphipathic chemical agents produced by living organisms as their secondary metabolites that can decrease surface and interfacial tension.1,2 Many environmental strains of microorganisms can produce biosurfactants since they are key agents for increasing the bioavailability of hydrophobic compounds, which can serve as a carbon source for microbial growth.1 Therefore, bacteria producing biosurfactants can access the hydrophobic phase and metabolize a variety of aliphatic hydrocarbons and polycyclic aromatic hydrocarbons (PAHs). Bacteria producing surfactants have also found many commercial applications, especially in remediation for the removal of hydrocarbon pollutants and heavy metals from the environment.2 Purified bacterial surfactants have been used to control pathogens in food products,3 as emulsion-stabilizing agents in the food industry,4 for drug delivery,5 as a potent and environmentally friendly biopesticide against plant pathogens,6 and in the cosmetic industry.7Existing approaches for screening of microorganisms producing biosurfactants include: i) spread assay tests, ii) emulsification tests, iii) hemolytic assays, and iv) surface tension measurements.1 However, these methods are time-consuming, low-throughput, non-quantitative, and characterized by relatively low selection accuracy. Typically, they necessitate microliter or milliliter volumes of samples, with one measurement taking from several seconds to a few minutes. Additionally, the cells of interest must be first enriched from a complex consortium of environmental microorganisms before screening the functional properties of bacterial cultures. Traditionally, strains of bacteria are enriched via cultivation on solid media (e.g., agar plates) or in liquid cultures. Among the above-mentioned methods, only hemolytic assays can be conducted concurrently with the cultivation of bacteria on agar plates, though the throughput is typically limited to a few hundred colonies per single plate. Multiple studies also reported significant differences in bacteria cultivability using liquid or solid media, with the reduced recovery of strains grown on agar.8 On the other hand, bulk liquid culture in rich media might result in a bias towards fast-growing strains overtaking the ones slowly growing9 that are potential biosurfactant producers.
To the best of our knowledge, there are no available high-throughput methods for screening of microorganisms producing biosurfactants that could overcome the above-mentioned limitations of enrichment in bulk liquid cultures and on agar plates. Droplet microfluidics has been recently applied for the successful selection of bacterial strains with higher taxonomic richness and larger representation of rare species,10,11 in comparison to standard methods of enrichment. However, these methods are mainly based on the growth of bacteria measured by cell concentration or optical readouts. Cell density measurements provide data on bacterial abundance in microdroplets but do not distinguish bacterial strains depending on their metabolic activity or functional properties. Optical-activated droplet sorting can be applied for measuring cell concentration – e.g., light scattering assay for detecting the growth of antibiotic-resistant bacteria12 or optical density assay for selecting slowly growing bacteria from complex gut microbiome isolate.11 Additionally, optical methods can be used for measuring selected aspects of the metabolic activity of living cells. Mahler et al. demonstrated the enrichment of microorganisms producing antibiotics via fluorometric assay to detect inhibition of the growth of reporter cells.13 In another example, a similar fluorescence-activated droplet sorting (FADS) technique was applied to screen microbes producing PET-degrading enzymes.14 However, the number of biochemical assays that can be measured by optical readouts in microdroplet format is still very limited.15 Consequently, the active sorting of droplets cannot easily be done for most functional screens of microbial cultures, including a selection of bacteria producing biosurfactants.
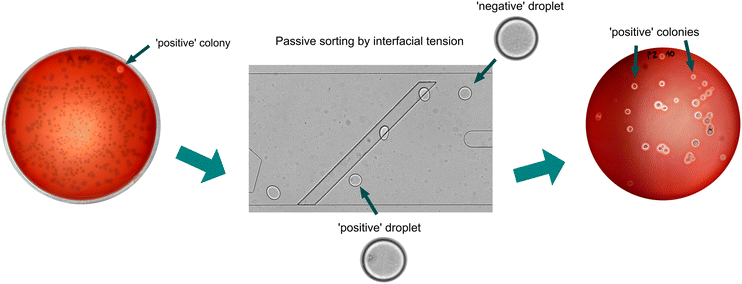
Here, we demonstrate a new application of passive droplet microfluidics for the enrichment of surfactant-producing bacteria. Our system is based on a method called “sorting by interfacial tension” (SIFT)16,17 – a label-free and passive technology that has been developed by the Abbyad group and so far used for several exciting demonstrations, such as i) isolation of single cells based on their metabolism,17 including cancer cells,18 and ii) sorting of droplets containing esterase enzymes.16 In another example, the group of Amar Basu demonstrated that migration of droplets in a surfactant gradient could be employed to sort droplets with and without bovine serum albumin based on differences in surface tension.19,20 Here, in our specific application, the passive sorting is based on the phenomenon that biosurfactants reduce the interfacial tension of each positive droplet containing bacteria producing biosurfactants (Fig. 1). We also investigated the impact of various parameters on the efficacy of droplet sorting, such as i) the concentration of water-soluble biosurfactants, ii) the volume of droplets, iii) oil flow rates needed for droplet sorting, iv) the time taken to cultivate bacteria in microdroplets, and v) the impact of continuous oxygenation of droplet culture. Finally, we validated our method via robust enrichment of the surfactant-producing bacterial strain from a mock consortium of bacteria.
Materials and methods
Bacteria cultivation
We used two strains of bacteria for the development and validation of the method: i) Bacillus sp. ANT_WA51 (ref. 21) strain for the generation of positive microdroplets, and ii) Escherichia coli BL21 strain as a negative control strain as it does not produce biosurfactants. Detailed protocols for bacteria cultivation and bulk surface tension measurements are provided in the ESI.†Fabrication of microfluidic devices
Microfluidic chips were fabricated using standard SU-8 photolithography and PDMS-based soft lithography – full protocols can be found in the ESI.†Droplet generation and cultivation of encapsulated bacteria
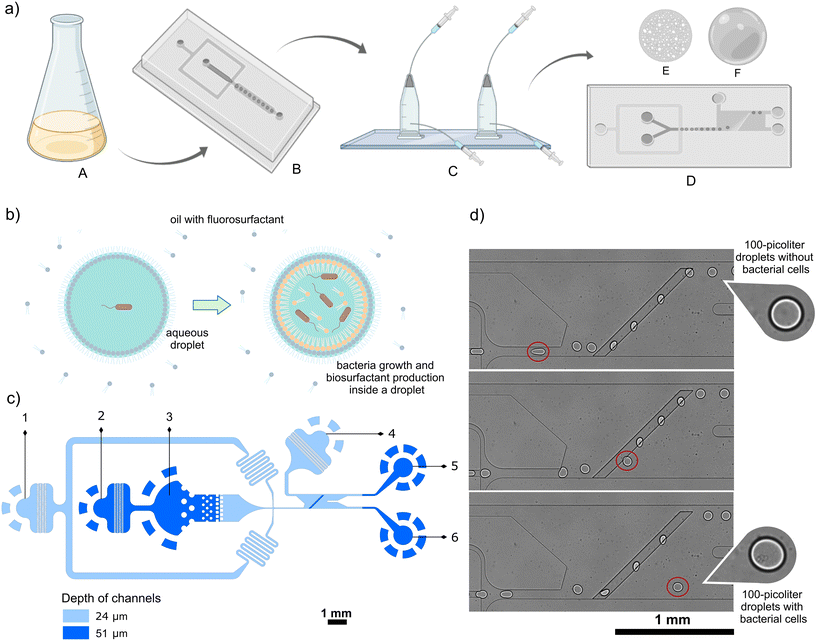
Bacterial cells were diluted in liquid LB medium to the appropriate concentration, and the suspension was transferred to sterile tubing and attached to a glass syringe filled with PBS buffer. Droplets were generated in a high-throughput microfluidic droplet flow-focusing generator with flow rates of 500 μL h−1 for the oil phase composed of Novec HFE-7500 (3M) with 2% 008-FluoroSurfactant (RAN Biotechnologies) and total 300 μL h−1 for cells in an aqueous medium (since our design had two aqueous inlets, we used two syringes providing bacteria suspension at a flow rate of 150 μL h−1 each). Around 200 μL of emulsion for each sample was collected in a dedicated chamber22 and incubated at room temperature for 24 hours, unless noted otherwise. For oxygenation of the emulsion, a peristaltic pump (Reglo ICC, Ismatec) was used, with dedicated Tygon tubing of 0.38 mm I.D, 0.9 mm O.D (Ismatec) that was next tightly connected to the PTFE tubing 0.4 mm I.D, 0.9 mm O.D (Bola Bohlender) and polyethylene tubing (Smiths Medical) of the incubation chamber (Fig. S2†). The system was closed by putting two sides of tubing into a 5 mL stock of 2% FluoroSurfactant in HFE-7500 to prevent air bubbles and to provide oxygen dissolved in the oil to the emulsion. A detailed protocol for oxygen supply to the emulsion is provided in the ESI.†Passive droplet sorting
Before the passive sorting process, gas-tight syringes (Hamilton) were filled with oil, attached to PTFE tubing, and placed tightly on syringe pumps (Nemesys, Cetoni). The outlet tubing of the droplet incubation chamber was connected to the microfluidic device for the emulsion transfer. The tubing for oil delivery from syringes onto the chip was also attached to oil inlets. Then, the pumps started to push oil and emulsion into the chip, and in most of the experiments, the emulsion was sorted passively with the following flow rates: 480 μL h−1 for spacing oil (2% FluoroSurfactant in HFE-7500), 20 μL h−1 for diluting oil (2% FluoroSurfactant in HFE-7500), 2500–3000 μL h−1 for sorting oil (pure HFE-7500), and 50–100 μL h−1 for the emulsion pushed with oil (2% FluoroSurfactant in HFE-7500). Videos were captured using a fast camera (AX-100, Photron) and an inverted microscope (CKX53, Olympus). Selected videos were analyzed using ImageJ software to determine the values of the displacement factor (a detailed description of image analysis can be found in the ESI†).Determination of enrichment factor
Two bacterial strains, Bacillus sp. ANT_WA51 and E. coli BL21, were separately cultured overnight, centrifuged (4000 rpm, 8 min) and rinsed with 0.9% NaCl saline solution, then mixed as a mock consortium in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, suspended in LB medium to achieve a concentration corresponding to approximately λ = 0.1, and encapsulated in 100 pL microdroplets. After overnight oxygenated cultivation, the emulsion was transferred to the microfluidic device, and around 20
100, suspended in LB medium to achieve a concentration corresponding to approximately λ = 0.1, and encapsulated in 100 pL microdroplets. After overnight oxygenated cultivation, the emulsion was transferred to the microfluidic device, and around 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets were sorted for 3 minutes to collect approximately 20 positive droplets, in sterile 1.5 mL tubes, in two replicates. The volume of oil in each sample was adjusted to 250 μL by flushing the used tubing with filtered HFE-7500. Next, 62.5 μL 1H,1H,2H,2H-perfluoro-1-octanol (PFO) (Alfa Aesar), and 200 μL of sterile 0.9% NaCl saline solution was added to the emulsion. The two-phase mixture was then vortexed for 90 seconds to break the emulsion and centrifuged for 60 seconds to let the aqueous and oil phases separate. Next, the aqueous phase was diluted by 10×, 100×, and 1000×, then plated on Petri dishes with LB agar (Miller) medium supplied with 10% (v/v) defibrinated horse blood (GRASO Biotech) for hemolytic assay,23 in which surfactants produced by bacteria cause beta-hemolysis visible as a transparent zone around a colony. Plates were incubated at room temperature for two days and then analyzed by visual counting of colonies. Colony-forming units were counted for each plate with a distinction between negative (no hemolysis zone around a colony) and positive (showing a hemolysis zone).
000 droplets were sorted for 3 minutes to collect approximately 20 positive droplets, in sterile 1.5 mL tubes, in two replicates. The volume of oil in each sample was adjusted to 250 μL by flushing the used tubing with filtered HFE-7500. Next, 62.5 μL 1H,1H,2H,2H-perfluoro-1-octanol (PFO) (Alfa Aesar), and 200 μL of sterile 0.9% NaCl saline solution was added to the emulsion. The two-phase mixture was then vortexed for 90 seconds to break the emulsion and centrifuged for 60 seconds to let the aqueous and oil phases separate. Next, the aqueous phase was diluted by 10×, 100×, and 1000×, then plated on Petri dishes with LB agar (Miller) medium supplied with 10% (v/v) defibrinated horse blood (GRASO Biotech) for hemolytic assay,23 in which surfactants produced by bacteria cause beta-hemolysis visible as a transparent zone around a colony. Plates were incubated at room temperature for two days and then analyzed by visual counting of colonies. Colony-forming units were counted for each plate with a distinction between negative (no hemolysis zone around a colony) and positive (showing a hemolysis zone).
Results and discussion
Development of the method for the selection of bacteria producing biosurfactants included i) designing and testing the tension-based passive droplet sorter, ii) optimization of cultivation inside picoliter droplets, iii) finding the best parameters for droplet sorting, and finally, iv) demonstration of enrichment of biosurfactant-producing bacteria from a mock microbial consortium.The design of the tension-based passive droplet sorter
Fig. 1c presents the layout of the microfluidic chip, which is a modified version of the SIFT system developed by the Abbyad group.16–18 The passive droplet sorter contains a reinjection chamber with inlets for diluting oil and emulsion, an inlet for spacing oil, a droplet spacing junction, a sorting chamber with a supply of sorting oil, and two outlets for droplet collection. The sorting process began with the reinjection of the droplet emulsion through the droplet inlet and the diluting oil in the reinjection chamber. Next, the spacing oil was introduced to accelerate droplets in the direction of the sorting module and space them evenly to avoid droplet collisions that negatively affect the sorting process. As the droplets entered the sorting module, they were squeezed into a pancake shape because the height of the sorting module chamber (24 μm) was smaller than the diameter of the 100 pL droplet, (equivalent to around 58 μm diameter for a spherical droplet of that volume, calculated with a dedicated formula provided in the ESI†). However, upon encountering a deeper rail (51 μm) inclined at a 45° angle to the flow direction, the droplets underwent a shape change to minimize their overall surface area. The rail then retains the confined droplets with a force proportional to the change in surface area. The positive droplets with lower interfacial tension were pushed off the rail early by sorting oil towards a positive outlet, unlike the negative droplets that flow along the rail till its peak. Positive droplets are directed to the separate outlet channel and can be collected off-chip in a tube for further culture upscaling and characterization of sorted bacteria. The droplet sorting can be triggered by changing the flow rate of sorting oil, as shown in Fig. S3.†The sorter design underwent several modifications, in comparison to previously published versions of interfacial tension-based sorters (Table S3†).16–18 The width of the sorting device was reduced from 1.5 mm to 0.75 mm, and the sorting rail was repositioned closer to the channel entering the chamber, which provides microdroplets to the sorting device. An additional diluting oil stream was introduced to the droplet reinjection chamber (inlet 2, Fig. 1c) to make an emulsion less densely packed and improve the subsequent spacing of droplets prior to sorting. Furthermore, a dilution of emulsion with oil also reduced unwanted fragmentation of droplets, especially positive ones containing high concentrations of biosurfactants. However, small satellite droplets remain visible in the videos depicting the sorting process (e.g. Videos S1 and S2†). We observed that satellites are primarily formed during the transport of droplets with high biosurfactant content, which exhibit lower stability. Typically, these satellite droplets are too small to be sorted on the rail and instead flow directly into the positive channel. However, given their predominant origin from positive droplets containing bacteria producing biosurfactants, the sorting efficiency remains unaffected in our application, as demonstrated in the enrichment experiment (Fig. 5). We achieved a droplet sorting frequency of up to 250, which is a higher value than 30 droplets per second in the SIFT systems presented before.16,17 The increased sorting frequency might be attributed to the larger interfacial tension between aqueous samples and continuous phase oil used in this study. This allowed faster flows of droplet emulsion while still achieving accurate sorting. The potential influence of other factors, such as the modified geometry of the sorting chamber and an increased depth of the rail, on the enhanced sorting throughput requires further investigations and systematic comparison of various geometries of the microfluidic modules.
Optimization of the flow parameters for the most efficient sorting
After several iterations of design and testing, we selected a final version of the sorting module and decided to optimize other parameters impacting the sorting efficiency, such as the droplet volume and flow rates of emulsion and oils. We chose a model organism, Bacillus sp. ANT_WA51, for the droplet-sorting optimization process. Bacillus sp. ANT_WA51 is a psychrophilic strain that produces a mixture of biosurfactants consisting mainly of surfactin and fengycin.21,24 Like pure surfactin, the mixture of biosurfactants produced by this strain can reduce the surface tension of growth media to 27–28 mN m−1.21,25 During the optimization process and determination of the sorting accuracy, we always used two populations of droplets: negative droplets containing pure LB medium, and positive droplets with encapsulated Bacillus sp. ANT_WA51 cells from overnight bulk liquid culture. Microdroplets of the same volumes were generated with dedicated flow-focusing chips and collected in a simple droplet chamber, allowing facile reinjection.22,26First, we determined the optimal volume of sorted droplets. The highest accuracy was observed for the 80–100 pL range of microdroplet volumes for tested sorter geometries. Microdroplets smaller than 80 pL were pushed off the rail too quickly, causing false positive errors and a loss of sorting specificity. In contrast, droplets with volumes larger than 100 pL were anchored too firmly to the rail and were prone to false negative sorting errors. Additionally, large droplets were less stable and prone to the fragmentation of spacing oil, and their sorting frequencies were generally lower. As a result of a series of experiments (Table S4†), microdroplets with a volume of 100 pL were chosen for further analysis and optimization.
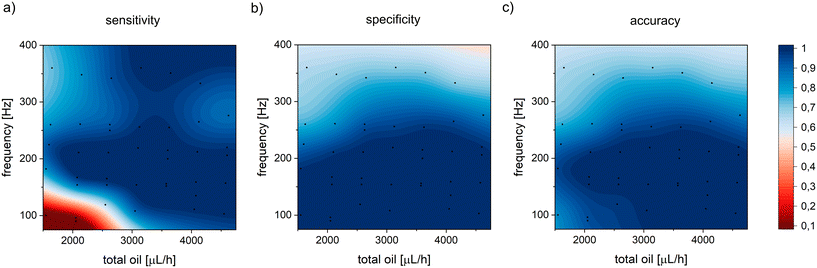
Next, we tested various flow rates of the spacing and sorting oils and determined the maximum throughputs for reliable sorting. For that purpose, we recorded and analyzed a series of videos with droplet-sorting experiments, and the numbers of true/false positive and true/false negative droplets were counted separately for each collection channel. As a true positive, we considered droplets with a bacterial colony collected in the positive channel, and false positives were empty droplets that flew to the positive channel. Accordingly, the empty droplets in the negative channel were regarded as true negative events, and the droplets with colonies in the negative channel were considered false negative events. Then, we calculated i) sensitivity as a ratio of true positive sorting events to the total number of droplets with colonies (sum of true positives and false negatives), and ii) specificity, which was the ratio of true negative events to the total number of empty droplets (sum of true negatives and false positives). Finally, we determined the accuracy as the ratio of true positive and true negative sorting events to the total number of droplets. We recorded sorting processes for a range of emulsion flow rates corresponding to various droplet frequencies of 100–350 droplets per second, and sorting oil flow rates of 1000–4000 μL h−1. The flow rates for diluting and spacing oil were fixed at 20 μL h−1 and 480 μL h−1, respectively. During the experiment, we injected negative (composed of pure LB media) and positive (with visible colonies) droplets at a ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to simulate the conditions of real screening experiments, where only a small fraction of droplets is sorted. Next, we analyzed the videos and manually counted negative and positive droplets sorted to both outlet channels. The highest accuracy, specificity, and sensitivity were achieved for total oil flow rates in the range of 2500–3000 μL h−1 and droplet frequencies of up to 200–250 Hz (Fig. 2 and Table S7†).
1 to simulate the conditions of real screening experiments, where only a small fraction of droplets is sorted. Next, we analyzed the videos and manually counted negative and positive droplets sorted to both outlet channels. The highest accuracy, specificity, and sensitivity were achieved for total oil flow rates in the range of 2500–3000 μL h−1 and droplet frequencies of up to 200–250 Hz (Fig. 2 and Table S7†).
Impact of biosurfactant concentration on sorting process
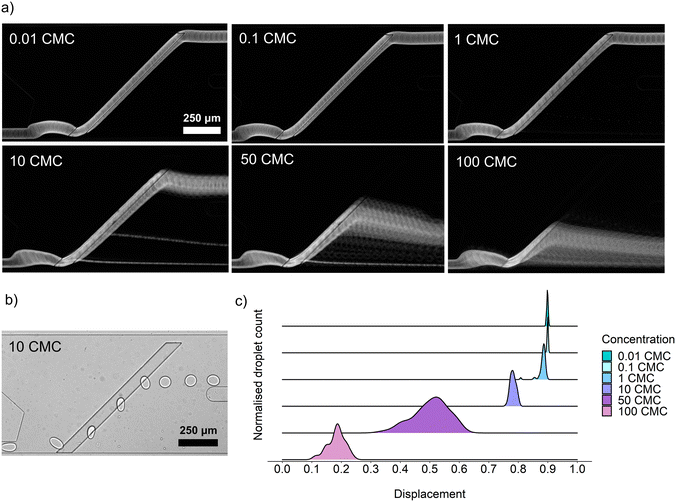
For the determination of a range of surfactant concentrations that can trigger the sorting of droplets, we chose surfactin, as it is a widely known lipopeptide biosurfactant produced by bacteria from the Bacillus genus and can efficiently reduce the surface tension of water solutions from around 72 mN m−1 down to 27 mN m−1.21,25 Another value characterizing each surfactant is the critical micelle concentration (CMC), which defines a concentration above which the surfactant molecules aggregate and form micelles. CMC values of surfactin vary in the literature from 10 mg L−1 to 20 mg L−1.27,28 In this assay, we used an intermediate CMC value of surfactin equal to 15.9 mg L−1 reported by Zou et al.25 Based on that value, a series of 100 pL droplet emulsions with 0.01 CMC, 0.1 CMC, 1 CMC, 10 CMC, 50 CMC, and 100 CMC were generated and sorted. Depending on the concentration of surfactin, microdroplets were detaching from the rail at various points, and the lateral distance between this detachment point and the initial position of the droplet was defined as a displacement value (Fig. 3c). Displacement equaled 0 when a droplet detached from the rail at the same width as the entry of the droplet supply channel. If the droplet flowed along the rail till the end, then the displacement value equaled 1. Displacement values lower than 0.6–0.7 usually resulted in the sorting of droplets to the positive outlet. Counterintuitively, the lowest surfactin concentration that allows for passive sorting turned out to be around 50 CMC and not 1 CMC, as might be expected from bulk measurement data available in the literature.29 This observation might suggest that sorting accuracy can not be directly predicted by using static surface tension measured in bulk conditions. The microdroplet emulsion, characterized by its elevated surface-to-volume ratio, demands a higher biosurfactant concentration to reach interface saturation and to trigger droplet sorting. However, quantitative assessment of this phenomenon is experimentally challenging and beyond the scope of this study.Optimization of the cultivation of bacteria in droplets
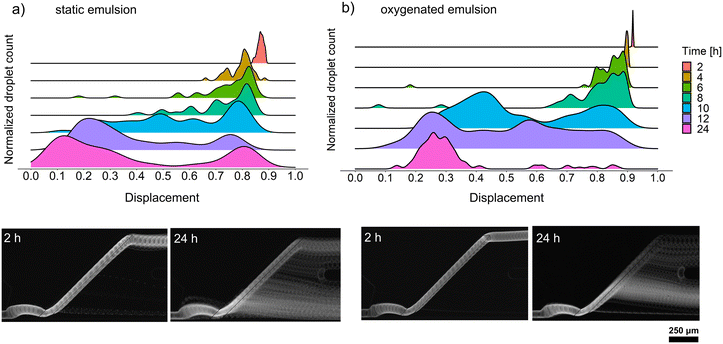
We decided to perform growth optimization using an emulsion consisting of only positive microdroplets to verify the efficiency of cultivation across the whole emulsion. For that purpose, we chose a cell concentration of λ = 5, corresponding to 5 cells per droplet on average, which, according to the Poisson distribution, implied a 99.3% probability that a randomly chosen microdroplet would contain at least one bacteria cell. However, our early attempts to passively sort droplets containing Bacillus sp. ANT_WA51 cells showed that not all droplets were sorted into the positive channel. Following the observations of Mahler and Zurek26,30 that oxygen availability in droplet emulsions is a key factor in the homogeneous growth of aerobic bacteria and the production of secondary metabolites, we decided to compare the growth of bacteria and biosurfactant production in static conditions, as well as with continuous oxygen supply. For that purpose, the kinetics of biosurfactant production by Bacillus sp. ANT_WA51 in a bulk culture supplied with oxygen by shaking the flask was first compared to an oxygen-limited bulk culture in a tightly closed tube, not subjected to shaking. The oxygen supply supported surfactant production measured by a decrease in the surface tension of culture supernatant.26,30 According to this bulk assay (Table S5†), Bacillus sp. ANT_WA51 started to produce biosurfactants after 4 hours of cultivation with an oxygen supply. However, it is important to note that bulk measurements were conducted at the liquid/air interface using the Wilhelmy plate method. These results should be viewed as indicators of the changes in interfacial tension that may be taking place for droplets in oil on the chip.To measure the biosurfactant production in microdroplet culture depending on oxygen supply, we incubated the emulsion containing Bacillus sp. ANT_WA51 cells using the setup with a peristaltic pump providing a constant oil flow to the emulsion.26 Two populations of droplets were incubated: the first one in a static condition and the second one under the 20 μL min−1 flow of the 2% RAN fluorosurfactant in HFE-7500 oil that provided oxygen to the emulsion. Next, we performed passive sorting for oxygenated and static emulsions every two hours to determine the displacement factor and an optimal incubation time for Bacillus sp. ANT_WA51. During this experiment, we used a sorting oil flow rate of 2500 μL h−1 and a droplet frequency of around 150 Hz. The optimal cultivation time for the Bacillus sp. ANT_WA51 strain was assessed for 24 hours, based on the displacement for sorting the oxygenated emulsion (Fig. 4). This result stands in contrast to the 4–8 hours needed to achieve minimal surface tension in an oxygenated bulk culture, as outlined in Table S5.† However, as illustrated in Fig. 3, the sorting of droplets is triggered for biosurfactant concentrations much higher than 1 CMC measured in bulk conditions. The oxygenated emulsion showed similar displacement values to the static one (Fig. 4). However, oxygen supply increased the homogeneity of trajectories of sorted microdroplets. As biosurfactants are complex compounds classified as secondary metabolites, their production usually occurs during the stationary phase.1,31 Low oxygen concentration slows down bacterial cell growth and might result in delayed onset of the stationary phase coupled with biosurfactant production. Limited oxygen supply in static conditions might additionally result in false negative events caused by heterogeneous growth in microaerobic conditions, which is consistent with the observations made by Mahler et al. and Zurek et al.26,30 for the cultivation of environmental strains and microbial growth coupled with protein expression, respectively. However, we also observed some variations in biosurfactant production within the oxygenated emulsion, visible at the 10- and 12-hour time points. This variability could potentially be attributed to the phenotypic differences of the cultures originating from single cells encapsulated in droplets at the beginning of the assay. Bacteria possess the ability to undergo differentiation, resulting in the emergence of subpopulations that may exhibit distinct phenotypes despite being genetically identical. An example of such differentiation into diverse cell types is exemplified by Bacillus subtilis species32 to which the strain Bacillus sp. ANT_WA51 belongs.24
Validation of the sorting accuracy by enrichment of positive strain Bacillus sp. ANT_WA51 from a mock microbial consortium
In the final stage of the project, we enriched the bacteria cultures originating from single cells encapsulated in droplets. Since a single-cell encapsulation follows Poisson distribution and most droplets are negative, we first had to demonstrate that biosurfactants do not diffuse among microdroplets during off-chip and long-term bacteria cultivation. Analysis of videos captured during the first attempts to passively sort approximately 2000 microdroplets containing diluted culture (λ = 0.1) provided information on each microdroplet's flow trajectory, as well as false positive events, which could be caused by the transfer of biosurfactant from positive microdroplets to negative one. These false positive events were not observed in the recorded videos, which means that the potential transfer of biosurfactants between droplets during bacteria cultivation is insufficient to impact sorting results.Next, we performed an enrichment experiment for the selection of rare positive cells from the excess bacteria that do not produce biosurfactants. For that purpose, two bacterial strains, Bacillus sp. ANT_WA51 and E. coli BL21, were mixed as a mock consortium in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and encapsulated in 100 pL microdroplets at λ = 0.1. After overnight-oxygenated cultivation, the emulsion was transferred to the microfluidic device, and around 20
100 and encapsulated in 100 pL microdroplets at λ = 0.1. After overnight-oxygenated cultivation, the emulsion was transferred to the microfluidic device, and around 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets were sorted for 3 minutes to collect around 20 positive droplets, which were plated on Petri dishes with LB agar medium supplied with 10% (v/v) defibrinated horse blood (see Method section). Surfactin can break down erythrocytes and free hemoglobin particles, which is visible as halos around positive bacterial colonies.23 Enrichment factors were calculated using two methods described previously by Baret et al. and Zinchenko et al.33,34 and were around 4250 and 119, respectively (Table S6†). Those high values prove that our method successfully enriches the bacteria producing biosurfactants.
000 droplets were sorted for 3 minutes to collect around 20 positive droplets, which were plated on Petri dishes with LB agar medium supplied with 10% (v/v) defibrinated horse blood (see Method section). Surfactin can break down erythrocytes and free hemoglobin particles, which is visible as halos around positive bacterial colonies.23 Enrichment factors were calculated using two methods described previously by Baret et al. and Zinchenko et al.33,34 and were around 4250 and 119, respectively (Table S6†). Those high values prove that our method successfully enriches the bacteria producing biosurfactants.
Conclusions
Passive droplet sorting is a promising approach characterized by the simplicity of the experimental setup, lower cost, and label-free detection of biological process outcomes. However, due to lower throughput and a limited array of possible assays, passive sorting is still less frequently used than optically-activated droplet sorting.33,35 Here, we expanded the application range of sorting based on the interfacial tension (SIFT) method (Table S3†), which has been used for passive sorting based on pH changes in microdroplets, including sorting of live and dead eukaryotic cells. We demonstrated that this approach could also be applied for environmental microbiology purposes to perform high-throughput screening for microorganisms secreting metabolites of interest that significantly decrease surface tension.To increase the throughput of droplet sorting, we experimentally tuned the microdroplet volume, and flow rates spacing and sorting oil to efficiently select droplets containing biosurfactant mixtures produced by the psychrophilic strain Bacillus sp. ANT_WA51. As a result, we reached passive sorting of up to 250 droplets per second. Our attempts to the additional increase the droplet sorting frequency in the 0.75 mm wide sorting module led to excessive sorting errors due to droplet collisions at the rail and fragmentation of droplets with spacing oil, as demonstrated in Video S3.† However, future studies focused on optimizing the depths and dimensions of sorting module elements, as well as the composition of the oil phase and droplet volume, might contribute to the further improvement of the sorting rate.
Apart from the characterization of physicochemical factors affecting passive sorting, we focused on adjusting the growth conditions of the bacteria microdroplet format. We demonstrated that additional oxygen supply via a flow of fluorocarbon oil through an emulsion with bacterial cells resulted in more homogeneous production of biosurfactant across microdroplets in the emulsion. We showed that single-bacteria cells encapsulated in microdroplets could efficiently grow and produce biosurfactants. However, only droplets with a concentration of biosurfactant exceeding the equivalent of 50 CMC (0.795 mg mL−1) of surfactin could be sorted. Therefore, we assumed that bacteria-producing biosurfactants are sorted only when the biosurfactant concentration in a microdroplet is higher than this value. However, this dependency might be complex and rely on various types and structures of biosurfactant molecules, possible interactions between biosurfactants and fluorosurfactants, and other factors influencing the dynamic interfacial tension in a flowing microdroplet. A better understanding of the physics and chemistry of the SIFT method is beyond the scope of this work, though it is worth evaluating in further research and will lead to additional improvement of the throughput of the system presented here.
The technology was validated by the enrichment of rare positive microbial cells. We obtained very high enrichment factors equaling 4250 and 119, depending on the calculation method used.33,34 Those high values mean that our passive droplet sorting method is highly effective in enriching rare bacterial cells of interest, providing that they grow in a liquid medium encapsulated in droplets.
One of the disadvantages of the current version of the system is the inability to select bacterial strains requiring induction for biosurfactant production, such as Pseudomonas species producing rhamnolipids.6 Such an induction step is usually performed by adding a hydrocarbon compound, which might be a challenging step in microdroplet format and will probably require the generation of multiple emulsions. In the next iterations of the system, we plan to introduce the induction of biosurfactant production inside microdroplets by e.g., encapsulating small hydrocarbon droplets inside aqueous growth media droplets or introducing water-soluble inductors.
Despite those limitations, we believe that many microorganisms constitutively producing biosurfactants, especially those growing slowly, can be efficiently selected with the current version of the system. The experimental strategy developed here will be used in the future to conduct screening campaigns from environmental samples to isolate novel strains of bacteria and other microorganisms producing biosurfactants.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Michał Styczyński and Łukasz Dziewit for sharing their Bacillus sp. ANT_WA51 strain. This research was funded by the TEAM-NET programme of the Foundation for Polish Science, project no. POIR.04.04.00-00-14E6/18-00 as a part of Measure 4.4 of the 2014–2020 Smart Growth Operational Programme, EU. Research infrastructure used in the project was co-funded by the “Excellence Initiative – Research University (2020–2026)” programme via Action I.4.2 “Fund for the Renovation and Development of Research Infrastructure.”References
- S. J. Varjani and V. N. Upasani, Critical Review on Biosurfactant Analysis, Purification and Characterization Using Rhamnolipid as a Model Biosurfactant, Bioresour. Technol., 2017, 232, 389–397, DOI:10.1016/j.biortech.2017.02.047.
- B. Liu, J. Liu, M. Ju, X. Li and Q. Yu, Purification and Characterization of Biosurfactant Produced by Bacillus Licheniformis Y-1 and Its Application in Remediation of Petroleum Contaminated Soil, Mar. Pollut. Bull., 2016, 107(1), 46–51, DOI:10.1016/j.marpolbul.2016.04.025.
- L. Magalhães and M. Nitschke, Antimicrobial Activity of Rhamnolipids against Listeria Monocytogenes and Their Synergistic Interaction with Nisin, Food Control, 2013, 29(1), 138–142, DOI:10.1016/j.foodcont.2012.06.009.
- G. S. Kiran, S. Priyadharsini, A. Sajayan, G. B. Priyadharsini, N. Poulose and J. Selvin, Production of Lipopeptide Biosurfactant by a Marine Nesterenkonia Sp. and Its Application in Food Industry, Front. microbiol., 2017, 8, 1138, DOI:10.3389/fmicb.2017.01138.
- T. R. Bjerk, P. Severino, S. Jain, C. Marques, A. M. Silva, T. Pashirova and E. B. Souto, Biosurfactants: Properties and Applications in Drug Delivery, Biotechnology and Ecotoxicology, Bioengineering, 2021, 8(8), 115, DOI:10.3390/bioengineering8080115.
- J. Chen, Q. Wu, Y. Hua, J. Chen, H. Zhang and H. Wang, Potential Applications of Biosurfactant Rhamnolipids in Agriculture and Biomedicine, Appl. Microbiol. Biotechnol., 2017, 101(23–24), 8309–8319, DOI:10.1007/s00253-017-8554-4.
- S. A. Adu, P. J. Naughton, R. Marchant and I. M. Banat, Microbial Biosurfactants in Cosmetic and Personal Skincare Pharmaceutical Formulations, Pharmaceutics, 2020, 12(11), 1099, DOI:10.3390/pharmaceutics12111099.
- K. Nosho, K. Yasuhara, Y. Ikehata, T. Mii, T. Ishige, S. Yajima, M. Hidaka, T. Ogawa and H. Masaki, Isolation of Colonization-Defective Escherichia Coli Mutants Reveals Critical Requirement for Fatty Acids in Bacterial Colony Formation, Microbiology, 2018, 164(9), 1122–1132, DOI:10.1099/mic.0.000673.
- W. H. Lewis, G. Tahon, P. Geesink, D. Z. Sousa and T. J. G. Ettema, Innovations to Culturing the Uncultured Microbial Majority, Nat. Rev. Microbiol., 2021, 19(4), 225–240, DOI:10.1038/s41579-020-00458-8.
- M. M. Villa, R. J. Bloom, J. D. Silverman, H. K. Durand, S. Jiang, A. Wu, E. P. Dallow, S. Huang, L. You and L. A. David, Interindividual Variation in Dietary Carbohydrate Metabolism by Gut Bacteria Revealed with Droplet Microfluidic Culture, mSystems, 2020, 5(3), e00864-19, DOI:10.1128/mSystems.00864-19.
- W. J. Watterson, M. Tanyeri, A. R. Watson, C. M. Cham, Y. Shan, E. B. Chang, A. M. Eren and S. Tay, Droplet-Based High-Throughput Cultivation for Accurate Screening of Antibiotic Resistant Gut Microbes, eLife, 2020, 9, e56998, DOI:10.7554/eLife.56998.
- X. Liu, R. E. Painter, K. Enesa, D. Holmes, G. Whyte, C. G. Garlisi, F. J. Monsma, M. Rehak, F. F. Craig and C. A. Smith, High-Throughput Screening of Antibiotic-Resistant Bacteria in Picodroplets, Lab Chip, 2016, 16(9), 1636–1643, 10.1039/C6LC00180G.
- L. Mahler, S. P. Niehs, K. Martin, T. Weber, K. Scherlach, C. Hertweck, M. Roth and M. A. Rosenbaum, Highly Parallelized Droplet Cultivation and Prioritization of Antibiotic Producers from Natural Microbial Communities, eLife, 2021, 10, e64774, DOI:10.7554/eLife.64774.
- Y. Qiao, R. Hu, D. Chen, L. Wang, Z. Wang, H. Yu, Y. Fu, C. Li, Z. Dong, Y.-X. Weng and W. Du, Fluorescence-Activated Droplet Sorting of PET Degrading Microorganisms, J. Hazard. Mater., 2022, 424, 127417, DOI:10.1016/j.jhazmat.2021.127417.
- S. Neun, P. J. Zurek, T. S. Kaminski and F. Hollfelder Ultrahigh Throughput Screening for Enzyme Function in Droplets, in Methods in Enzymology, Elsevier, 2020, vol. 643, pp. 317–343, DOI:10.1016/bs.mie.2020.06.002.
- D. G. Horvath, S. Braza, T. Moore, C. W. Pan, L. Zhu, O. S. Pak and P. Abbyad, Sorting by Interfacial Tension (SIFT): Label-Free Enzyme Sorting Using Droplet Microfluidics, Anal. Chim. Acta, 2019, 1089, 108–114 CrossRef CAS.
- C. W. Pan, D. G. Horvath, S. Braza, T. Moore, A. Lynch, C. Feit and P. Abbyad, Sorting by Interfacial Tension (SIFT): Label-Free Selection of Live Cells Based on Single-Cell Metabolism, Lab Chip, 2019, 19(8), 1344–1351 RSC.
- C. Zielke, C. W. Pan, A. J. Gutierrez Ramirez, C. Feit, C. Dobson, C. Davidson, B. Sandel and P. Abbyad, Microfluidic Platform for the Isolation of Cancer-Cell Subpopulations Based on Single-Cell Glycolysis, Anal. Chem., 2020, 92(10), 6949–6957, DOI:10.1021/acs.analchem.9b05738.
- G. K. Kurup and A. S. Basu, Passive, Label-Free Droplet Sorting by Chemical Composition Using Tensiophoresis, Micro Total Anal. Syst., 2012, 2012, 76–78 Search PubMed.
- G. K. Kurup and A. S. Basu, Size Based Droplet Sorting with Wide Tuning Range Using Tensiophoresis, Micro Total Anal. Syst., 2013, 2013, 1344–1346 Search PubMed.
- M. Styczynski, G. Biegniewski, P. Decewicz, B. Rewerski, K. Debiec-Andrzejewska and L. Dziewit, Application of Psychrotolerant Antarctic Bacteria and Their Metabolites as Efficient Plant Growth Promoting Agents, Front. Bioeng. Biotechnol., 2022, 10, 772891, DOI:10.3389/fbioe.2022.772891.
- S. Neun, T. S. Kaminski and F. Hollfelder, Single-Cell Activity Screening in Microfluidic Droplets, in Methods in Enzymology, Elsevier, 2019, vol. 628, pp. 95–112, DOI:10.1016/bs.mie.2019.07.009.
- S. Joy, P. K. S. M. Rahman and S. Sharma, Biosurfactant Production and Concomitant Hydrocarbon Degradation Potentials of Bacteria Isolated from Extreme and Hydrocarbon Contaminated Environments, Chem. Eng. J., 2017, 317, 232–241, DOI:10.1016/j.cej.2017.02.054.
- T. Krucoń, Z. Ruszkowska, W. Pilecka, A. Szych and Ł. Drewniak, Bioprospecting of the Antarctic Bacillus Subtilis Strain for Potential Application in Leaching Hydrocarbons and Trace Elements from Contaminated Environments Based on Functional and Genomic Analysis, Environ. Res., 2023, 115785, DOI:10.1016/j.envres.2023.115785.
- A. Zou, J. Liu, V. M. Garamus, Y. Yang, R. Willumeit and B. Mu, Micellization Activity of the Natural Lipopeptide [Glu 1, Asp 5] Surfactin-C15 in Aqueous Solution, J. Phys. Chem. B, 2010, 114(8), 2712–2718, DOI:10.1021/jp908675s.
- P. J. Zurek, R. Hours, U. Schell, A. Pushpanath and F. Hollfelder, Growth Amplification in Ultrahigh-Throughput Microdroplet Screening Increases Sensitivity of Clonal Enzyme Assays and Minimizes Phenotypic Variation, Lab Chip, 2021, 21(1), 163–173, 10.1039/D0LC00830C.
- M. Deleu, H. Razafindralambo, Y. Popineau, P. Jacques, P. Thonart and M. Paquot, Interfacial and Emulsifying Properties of Lipopeptides from Bacillus Subtilis, Colloids Surf., A, 1999, 152(1–2), 3–10, DOI:10.1016/S0927-7757(98)00627-X.
- R. Sen and T. Swaminathan, Characterization of Concentration and Purification Parameters and Operating Conditions for the Small-Scale Recovery of Surfactin, Process Biochem., 2005, 40(9), 2953–2958, DOI:10.1016/j.procbio.2005.01.014.
- T. Janek, E. J. Gudiña, X. Połomska, P. Biniarz, D. Jama, L. R. Rodrigues, W. Rymowicz and Z. Lazar, Sustainable Surfactin Production by Bacillus Subtilis Using Crude Glycerol from Different Wastes, Molecules, 2021, 26(12), 3488, DOI:10.3390/molecules26123488.
- L. Mahler, M. Tovar, T. Weber, S. Brandes, M. M. Rudolph, J. Ehgartner, T. Mayr, M. T. Figge, M. Roth and E. Zang, Enhanced and Homogeneous Oxygen Availability during Incubation of Microfluidic Droplets, RSC Adv., 2015, 5(123), 101871–101878, 10.1039/C5RA20118G.
- T. Danevčič, A. Dragoš, M. Spacapan, P. Stefanic, I. Dogsa and I. Mandic-Mulec, Surfactin Facilitates Horizontal Gene Transfer in Bacillus Subtilis, Front. microbiol., 2021, 12, 657407, DOI:10.3389/fmicb.2021.657407.
- F. B. Rahman, Molecular Genetics of Surfactin and Its Effects on Different Sub-Populations of Bacillus Subtilis, Biotechnol. Rep., 2021, 32, e00686, DOI:10.1016/j.btre.2021.e00686.
- J.-C. Baret, O. J. Miller, V. Taly, M. Ryckelynck, A. El-Harrak, L. Frenz, C. Rick, M. L. Samuels, J. B. Hutchison, J. J. Agresti, D. R. Link, D. A. Weitz and A. D. Griffiths, Fluorescence-Activated Droplet Sorting (FADS): Efficient Microfluidic Cell Sorting Based on Enzymatic Activity, Lab Chip, 2009, 9(13), 1850, 10.1039/b902504a.
- A. Zinchenko, S. R. A. Devenish, B. Kintses, P.-Y. Colin, M. Fischlechner and F. Hollfelder, One in a Million: Flow Cytometric Sorting of Single Cell-Lysate Assays in Monodisperse Picolitre Double Emulsion Droplets for Directed Evolution, Anal. Chem., 2014, 86(5), 2526–2533, DOI:10.1021/ac403585p.
- E. J. Medcalf, M. Gantz, T. S. Kaminski and F. Hollfelder, Ultra-High-Throughput Absorbance-Activated Droplet Sorting for Enzyme Screening at Kilohertz Frequencies, Anal. Chem., 2023, 95(10), 4597–4604, DOI:10.1021/acs.analchem.2c04144.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed protocols for the photolithographic microfabrication of molds of microfluidic devices and assembly of chambers for droplet incubation, a protocol for the emulsion oxygenation, sorting parameters with comparison to the SIFT method, tables with values of sorting sensitivity, specificity, and accuracy calculated for chosen droplet volumes, results of surface tension measurements, and video captions. CAD file with microfluidic chip designs (.dxf). Video S1 of passive sorting of droplets with diluted Bacillus sp. ANT_WA51 bulk culture (AVI). Video S2 of passive sorting during enrichment of 2-strain mix (AVI). Video S3 of passive sorting with high droplet frequency in a 0.75-mm-wide chamber. Collisions of and break-up of droplets resulted in sorting errors (AVI). See DOI: https://doi.org/10.1039/d3lc00656e |
| This journal is © The Royal Society of Chemistry 2024 |