Open Access Article
Open Access ArticleMicroplastic emerging pollutants – impact on microbiological diversity, diarrhea, antibiotic resistance, and bioremediation
Karupanagounder Thangaraj
Uthra
a,
Vellapandian
Chitra
b,
Narayanasamy
Damodharan
c,
Anitha
Devadoss
d,
Moritz
Kuehnel
 ef,
Antonio Jose
Exposito
ef,
Antonio Jose
Exposito
 g,
Sanjay
Nagarajan
g,
Sudhagar
Pitchaimuthu
g,
Sanjay
Nagarajan
g,
Sudhagar
Pitchaimuthu
 *h and
Gururaja Perumal
Pazhani
*a
*h and
Gururaja Perumal
Pazhani
*a
aDepartment of Pharmaceutical Chemistry, SRM College of Pharmacy, SRM Institute of Science and Technology, Kattankulattur, Chennai 603 203, Tamil Nadu, India. E-mail: pazhanigp@gmail.com
bDepartment of Pharmacology, SRM College of Pharmacy, SRM Institute of Science and Technology, Kattankulattur, Chennai 603 203, Tamil Nadu, India
cDepartment of Pharmaceutics, SRM College of Pharmacy, SRM Institute of Science and Technology, Kattankulattur, Chennai 603 203, Tamil Nadu, India
dInstitute of Biological Chemistry, Biophysics and Bioengineering, School of Engineering & Physical Sciences, Heriot-Watt University, Edinburgh, UK
eDepartment of Chemistry, Faculty of Science and Engineering, Swansea University, Singleton Park, SA2 8PP, Swansea, Wales, UK
fFraunhofer Institute for Wind Energy Systems IWES, Am Haupttor 4310, 06237 Leuna, Germany
gDepartment of Chemical Engineering, University of Bath, Bath BA2 7AY, UK
hResearch Centre for Carbon Solutions, Institute of Mechanical, Processing and Energy Engineering, School of Engineering and Physical Sciences, Heriot-Watt University, Edinburgh, UK. E-mail: s.pitchaimuthu@hw.ac.uk
First published on 18th September 2023
Abstract
Advanced economic development and technologies cause worldwide plastic waste to increase many folds, leaving policymakers with the dilemma of managing it. Synthetic solid particles or polymeric matrices of plastics with diverse shapes and sizes are the primary concern of environmental pollution of the marine ecosystem, freshwater, agriculture fields, atmosphere, food, drinking water, and other remote locations. Researchers demonstrated microplastics (MPs) as multifaceted stressors in the ecosystem, carrying toxic chemicals and vectors of transport, and described the implications of these hazardous chemicals on human health. MPs in the environment can adsorb organic, nitrogenous substances and other minerals. This complex system may promote microbial growth and aggregation. Continuous contact of microbes with MPs changes the internal arrangement of ions and atoms, alternating physio-chemical properties and becoming hydrophobic. These properties allow specific bacterial growth on MPs and promote bacterial resistance and transfer of resistance genes. MPs aged by ultra-violet light, temperature, and chemicals increase bacterial adsorption and antibiotic-resistance gene transfer synergistically. MPs are mitigated in the environment by aggregation of microbes, which leads to aging and loss of the crystalline structure of microplastic due to the release of enzymes that cause oxidation, demethylation and desertification, and hydrolysis of MPs. Aerobic conditions are preferred to degrade MPs in different environmental conditions for large-scale degradation of MPs. However, anaerobic degradation requires controlled conditions and specialized equipment. The use of a consortium of bacteria increases biodegradation efficiency. Among the microorganisms, fungi were the most effective at detoxicating xenobiotics in the environment due to their adaptability and ability to tolerate diverse conditions. This critical review analyses microplastic-induced microbial diversity and microbial adaptations to it. Furthermore, it describes MP's role in the cause of diarrhea, antimicrobial resistance, and spread. The potential use of bioremediation methods and pathways for eliminating MPs like phthalates and bisphenol from ecosystems is discussed in detail. Finally, suggestions are put forward for controlling and removing MPs from the environment.
Environmental significanceIn today's world, plastic is everywhere because of its extensive use and durability. A total of 7800 million tonnes of plastics were manufactured between 1950 and 2015, with half of that production occurring between 2002 and 2015. As a result, plastic pollution has a significant and wide-ranging influence on people's health, the economy, tourism, and beach aesthetics. Microplastics (MPs) in the coastal and marine environment significantly harm the trophic levels of the food web, including bivalves, crustaceans, fish, and mammals. It can penetrate the intestinal wall or clog the intestines, which can mechanically harm an organism's digestive tract and reduce its ability to absorb nutrients. It decreases the energy reserves and feeding activity of marine worms. The power of microplastic absorbs toxic chemicals, which may then be consumed by aquatic organisms and causes severe health hazards to them. Humans are exposed to MPs in several ways, including seafood, drinking water, inhalation, and dermatological contact. MPs ingested through inhalation of particles breathed will either stay confined in the lungs or reach the digestive system through mucociliary clearance. MP in the respiratory tract leads to inflammation and respiratory discomfort in breaking. However, the knowledge that MPs exist in human lung tissues and their association with health implications provides new research avenue. The fate and bioaccumulation of ingested plastics remain unknown within the digestive tract of animals. There is a significant knowledge gap in understanding ubiquitous organic environmental pollutants like micro/nanoplastics in biological systems. MPs are believed to enter the intestine through food and water, which is also a carrier for pathogenic bacteria. This review will contribute to the understanding of MPs role in microbial adaptations, diarrhea, antimicrobial resistance, and probable mechanism of mitigation of MPs by bioremediation methods. Especially, the pathways for eliminating MPs like phthalates and bisphenol from ecosystems is discussed in detail alongside its advantages and disadvantages. Finally, suggestions are put forward for controlling and removing MPs from the environment. |
Introduction
In today's world, plastic is everywhere because of its extensive use and durability.1 A total of 7800 million tonnes of plastics were manufactured between 1950 and 2015, with half of that production occurring between 2002 and 2015.2 Due to the COVID-19 epidemic, its manufacturing and use have expanded significantly over the previous two decades, reaching an astounding level. During the COVID-19 epidemic, 1.6 million tonnes of plastic garbage were produced per day, primarily due to the increased production of disposable personal protective equipment. Which equates to 75 kg of plastic waste produced annually per person.3 Environmental MPs are classified into two categories primary, artificially generated micro-sized polymers made through extrusion or grinding, and secondary, a fragmentation of larger plastics by physical, biological, and chemical processes.4 Abiotic and biotic processes cause plastics to age and change in size when submerged in the marine environment. Mechanical abrasion and ultraviolet radiation exposure under simulated coastal conditions produced millions of secondary MPs.5 MPs (MP) are found in marine and freshwater habitats, such as beaches, seawater, marine sediments, surface water, and silt from lakes and rivers.6Estuaries and rivers are the sources of most MP in the ocean and coastal waters.1 In addition, current research has conclusively demonstrated that soils accumulate more MPs than aquatic habitats.3 Due to poor degradation rates and inappropriate usage and disposal, the amount of plastic in the environment is growing yearly.6 A total of 368 million tonnes of plastic were produced in 2019 due to the industrialization of several countries. About 40 million tonnes of plastic are still estimated to enter the environment each year, of which 11 million tonnes are the macro- and microplastic (MP) debris that ends up in the ocean.7 Due to increased shipping activities, Arctic surface waters and sea ice have been found with MPs.8 The existence of MPs in Arctic surface is an alarming situation of contamination in the environment and the persistence of MPs worldwide. MPs were found in the water, sediments, and biota of Asian freshwater ecosystems, which are considered a “hot spot” for plastic manufacture. Asia is home to the top 20 polluting rivers, which provide more than two-thirds of the yearly global inflow. Global modelling studies rank the Southeast Asian rivers are the largest source of plastics entering the ocean.9 India's production, imports, and exports of plastics have dramatically expanded during the past three decades to satisfy the nation's rapidly growing population. Plastic pollution has a significant and wide-ranging influence on people's health, the economy, tourism, and beach aesthetics. MPs in the coastal and marine environment significantly harm the trophic levels of the food web, including bivalves, crustaceans, fish, and mammals.10 It can penetrate the intestinal wall or clog the intestines, which can mechanically harm an organism's digestive tract and reduce its ability to absorb nutrients. It decreases the energy reserves and feeding activity of marine worms. The power of microplastic absorbs toxic chemicals, which may then be consumed by aquatic organisms and causes severe health hazards to them.10 Humans are exposed to MPs in several ways, including seafood, drinking water, inhalation, and dermatological contact. MPs ingested through inhalation of particles breathed will either stay confined in the lungs or reach the digestive system through mucociliary clearance.11 MP in the respiratory tract leads to inflammation and respiratory discomfort in breaking. However, the knowledge that MPs exist in human lung tissues and their association with health implications provides new research avenues.12 The residues of polystyrene (PS) and polyvinyl chloride (PVC) are commonly acquired micro- and nano-plastics (MNPLs) in human implants via day-to-day activities like ingestion, inhalation, and dermal exposure. The long-term persistence of these MNPLs in different organs and tissues leads to cancer development. A recent review emphasized that MNPLs have the potential to induce fibrosis, inflammation, and genotoxicity.13,14 The fate of MPs that reach the digestive system through oral and mucociliary clearance potentially impacts the gastrointestinal tract. Experimental demonstration of in vitro and in vivo in mammals provided evidence for MPs that affects the epithelial barrier, gut microbial imbalance, catabolic disorders, and endocrine disturbance. MP's distribution and adverse effect in GIT and organs depend upon MPs' nature, size, and physiochemical properties.15 MPs may persist in the colon and be cleared through urine and feces.
A human colonoscopy study demonstrated the presence of filaments or fibers in the colon, mainly polycarbonate (90%), polyamide, and polypropylene.16 The human excretion of MPs through urine has not been reported yet. Still, a recent rat animal model experiment evidenced that both nano and MPs that entered the blood circulation were excreted through urine with size ranges between 100 nm and 3 μm.17 Although MPs were detected in faeces and urine, the role of the cause of diarrhoea is yet not clearly understood. The smaller-sized MP distributed in the environment is unsuitable for recycling disposal using procedures designed for big plastic.18 Several conventional methods can remove plastics, including thermo-oxidative degradation, ultrafiltration, wet-air oxidation, reverse osmosis, coagulation, thermal degradation, biodegradation, ozonation, sonolysis, photocatalysis, and the Fenton process.19 Among all these methods, biodegradation is green technology for removing MPs. This process involves a variety of biological agents, including higher eukaryotes, fungi, and bacteria. These microorganisms can interact with MPs, aggregate on the MPs, and alter MPs' surface morphology and crystal lattice, leading to degradation. Often, extracellular enzymes produced by microorganisms can convert complex organic matter into simpler forms that support growth bacteria as a carbon source. Several pure cultures and microbial consortia have been successfully demonstrated to degrade MPs. Depending on the organisms, molecular mass and size of MPs and other environmental attributes greatly influence the microbes-assisted transformation.20,21 Microplastic recently got tremendous attention from researchers of biological and ecological domains. Although the term microplastic is debatable,22 the environmental impact of microplastics has been extensively studied.23 There is growing scientific evidence of microplastics association with various human illnesses. However, microplastics are not yet wholly elucidated on human health with the clinical specimen. It is genuinely unknown that the persistence of ingested plastics remains within the digestive tract of animals. There is a significant research gap in understanding ubiquitous organic environmental pollutants like micro/nanoplastics in biological systems. Microplastics are thought to enter the intestine through food and water, which is also a carrier for pathogenic bacteria. This review will contribute to the understanding of MPs role in microbial adaptations, diarrhea, antimicrobial spread, and probable mechanism of mitigation of MPs by microbes, which will outline the new avenues for possible research to mitigate the impact of MPs on human health.
Microbial diversity and adaptation with MPs
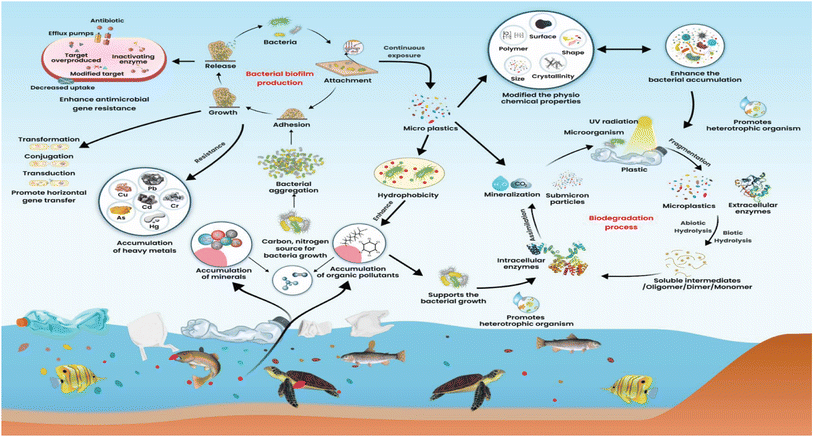
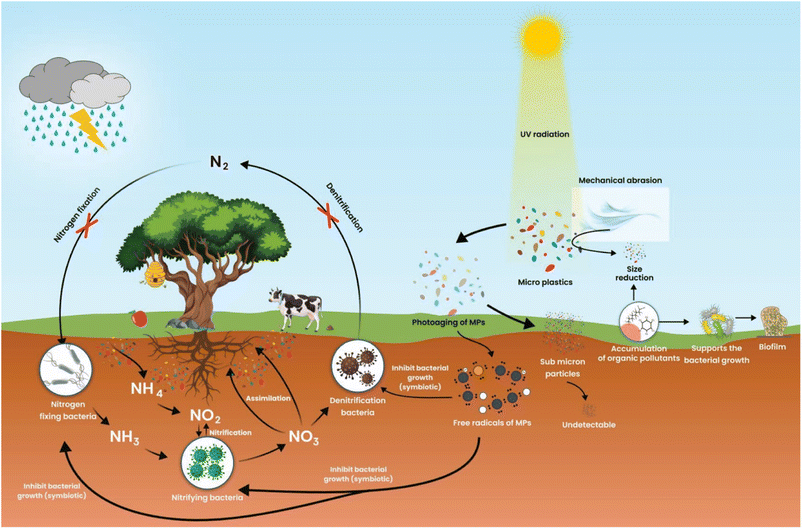
In low-income and developing countries, microbes pollute the environment. The major causes of microbial pollution are industrial effluents, sewage, and natural microbial communities. Microbes in this background can bioengineer themselves to fight against various chemicals, antimicrobials, and other environmental and physical factors. They join together in biofilm, providing the greatest strength to microbes to fight against several antibiotics and chemicals. Microbes can easily survive in the environment by consuming dissolved natural carbon concentrations of 30.7 μmol L−1. However, microorganisms will adapt, evolve and support the degradation of plastics. Microorganisms offer a wide variety of pathways to degrade plastics to plastic metabolites. The potential adaptations of bacteria on the MPs are a crucial feature of microorganism fitness, growth, and cell reproduction in different niches.24 However, scientific evidence has yet to reveal a link between MPs and the development of microorganisms.As a result of marine ecosystems being biologically more diverse than their equivalent terrestrial ecosystems, microorganisms are found in greater abundance in marine ecosystems. The microorganisms growing on microplastic are opportunistic organisms. Macro and MPs are the primary pollutants present in the environment of urban and semi-urban surface water. These plastics float on the water and hold several organic and other filthy matter together. MPs are chemically polymeric and remain inert on environmental surfaces in the form of natural organic aggregates and seston. These can adsorb nutrients from the surroundings and provide nutrients such as carbon, nitrogen, and other elements required for bacterial growth (Fig. 1).25 Different MPs can significantly affect nitrogen cycling processes in sediments, which may serve as organic carbon substrates for microbial communities (Fig. 2). Chemical processes such as nitrification and denitrification of soils cause polyurethane foam (PUF) or polylactic acid (PLA) to become a by-product of organic carbon substrates needed for microbial growth. Still, PVC-fortified soil inhibited both processes.26 Heavy metals with MPs in the soil can affect soil fertility, enzymes, catabolic functions, microbial diversity, total organic carbon (TOC), total nitrogen (TN), pH value, and biodegradation xenobiotics.27 MPs size and shape indirectly exacerbated microbial nutrient availability, metabolism and nutrient cycling, etc.28 The organic aggregates of MPs on the soil can exert selective pressure on the microbes in the vicinity of aggregates that allow the microbes to survive, adopt a unique substrate for colonization and support evolutionary changes.29
A high concentration of MPs (>10%) present in soil could compete with sediment microorganisms of niches and also be able to migrate downward into the soil of a certain distance.30 It supports the dominant growth of Chloroflexi, Proteobacteria, Bacteroidota, Firmicutes, and Actinobacteria.31 MPs cause an abundance of Proteobacteria and Bacteroidota in the soil. Low-density polyethylene MPs with concentration ranges of 2–7% could affect the Earth's bacterial diversity. In addition, it enhances the genes that encode nitrogen cyclings, such as nifH, AOBamoA, and nirK, in different phages.32 The constant contact of microbes with MPs changes properties such as crystallinity and becomes hydrophobic. These properties of MPs allow heterotrophic organisms such as cyanobacteria and Basidiomycota to grow on them. It increases the abundance of microplastic-degrading bacteria,33 promotes microbiota composition changes, uptake of isotopic elements, and alters growth and reproduction.34
Low-density polyethylene microplastic concentration ranges from 2–7%, affecting the soil bacterial diversity. In addition, it enhances the genes that encode nitrogen cyclings, such as nifH, AOBamoA, and nirK, in different phages.32 Wastewater treatment plant (WWTP) effluents have been considered pathways for MPs entry into aquatic environments. This microplastic favours the attachment of specific bacteria and organisms that could carry antimicrobial resistance genes that provide unique tolerance to selective antibiotics and environmental pressures, thereby increasing abundance and spread.
MPs associated diarrheal pathogens
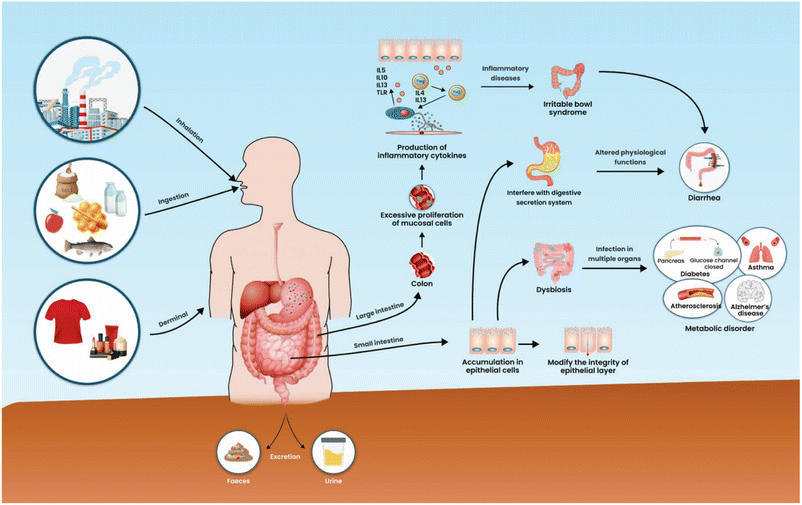
Inadvertent ingestion of MPs by humans through the oral route of contaminated foods and water leads to various MPs in the stool (Fig. 3).35 The ingested MPs could accumulate in the epithelial cells of the intestine, and the persistence of MPs in the intestine needs more scientific evidence. MPs are widespread environmental pollutants found in accumulation in various tissues of human beings, yet the risk of MPs in the occurrence of diarrheal disease is unclear. In mice models, microplastic exposure had a limited impact on the jejunum and liver. However, in the colon, the over-proliferation of mucosal cells leads to increased secretion of inflammatory markers such as IL-1β and IL-6 levels, increase in TNF-α levels, colitis with bloody diarrhea, and overactivation of the Notch signalling pathway in intestinal organoids.36 Secretion of these inflammatory markers could affect the jejunum and liver. Inflammatory markers are vital regulators of progressing inflammatory bowel disease.37 However, microplastic presence could injure the intestinal epithelial cell and also causes dysbiosis that leads to infections in multiple systems.38 Recently, MPs can interfere with the secretion system and affected normal physiological functions.39 Intestinal epithelial cell disturbance and alteration of normal physiological function in the intestine lead to a potential risk of inflammatory intestinal disease.40 Nanoplastics (NPs) of polyvinyl chloride (PVC) size ranges between 50–310 nm exposure to human monocytes and monocyte-derived dendritic cells provoked immune cells to secrete cytokines which leads to a progression of inflammation.41 Chronic diarrhea is closely related to inflammatory disease, and the cause of inflammation in the intestine may be associated with bacteria or xenobiotics.The mathematical models revealed the relationship between chronic diarrhea and urinary phthalate metabolite in adult patients from the USA. In the multivariate logistic regression analysis, urinary phthalate metabolite concentrations correlated with a risk of chronic diarrhea, and phthalates promote chronic diarrhea, particularly in adults.42 Several confounding factors were identified, such as complex pathophysiological conditions, intestinal inflammation, and obesity.42 Microplastic was detected in the feces of patients with inflammatory bowel diseases (IBD) with a concentration of 41.8 items per g dm than healthy people at 28.0 items per g.42 Polyethylene terephthalate and polyamide were prominent MPs detected among the several MPs present in the fecal samples. The contamination in humans may be plastic packaging of drinking water, food, and dust exposure.43 However, the correlation between MP ingestion, resident time of MPs in the human body, the onset of IBD, recurrence, and the total amount of MPs present in the stool specimens are yet to be established.43,44
Human pathogenic microbes in the plastisphere
The environmental biofilm of bacteria on plastic debris is called the “plastisphere”. It has a remarkable tendency to recruit a unique species of microorganism to grow in the aquatic environment than the sea ecosystem and the potential enough to create diverse microbial community populations in different ecosystems and cause microbial invasion. MPs significantly impact microbial catabolism in ecosystems, including carbon, nitrogen, sulfur cycles, compound degradation, and pathogenicity.45 A close relationship exists between environmental microorganisms on the plastisphere (macroplastics to nano-size plastics) and natural health. Several organisms have been found on plastics' surfaces, including pathogenic bacteria, which implies plastics may act as carriers of microbes that lead to the spread of diseases.46 However, the specific microbial populations on the plastisphere depend on several factors, such as environment, geographical regions, season, nutrient availability, and types of plastics.47 When MPs are ingested into animals, it can alter the structure of gut microbiota, create an imbalance among the commensal bacterial populations, and supports the growth of pathogens. By cellular internalization, it can also traverse the gastrointestinal tract into the tissue and cause serious health problems.48However, different types of MPs, like polyethylene and polystyrene, support the growth of diverse populations, and the dominance of certain species depends on the exposure duration to MPs. Virgin MPs tested in zebrafish (Danio rerio) models induced changes in metabolome and gut microbiota. As a result of these changes, zebrafish gut metabolites such as 8-amino-7-oxononanoate, cholesterol fatty acid ester, phosphatidylinositol, and n-triacontanol were altered. The gut microbiota, such as Fusobacteria and Proteobacteria were allowed to dominate over Firmicutes.49 A PS-MP with a diameter of 32–40 meters and a concentration of 100–1000 g L−1 has been observed to increase the size of goblet cells in juvenile guppies (Poecilia reticulata). Metabolic enzyme activity was also reduced, including trypsin, chymotrypsin, amylase, and lipase. In addition, the immune response such as TNF-α, IFN-γ, TLR4, and IL-6 elicited by MPs after 28 days of exposure.50 Exposure of polyvinyl chloride with a size of 2 μm and concentration of 100 mg kg−1 exposed intestine of rat models showed to cause changes in gut histology, biomarkers, and expression of mRNA expression levels of colonic mucus secretions.51 Hence, MPs can alter chemical compositions like bile salts and small and long-chain fatty acids, which are among the components that induce microbial diversity in the GIT. Humans are continuously exposed to MPs by inhaling and ingesting them through oral. Infants are prone to get exposure to MPs through toys.
A study of the impact of polyethylene MPs on infants' intestines was conducted using stimulated artificial colons coupled with co-cultures of epithelial and mucus cells. The decreased butyrate production and changes in volatile organic compound availability in the intestine are harmful to pathobionts of Dethiosulfovibrionaceae and Enterobacteriaceae.52 The Vibrio species are a marine organism that causes infection in fish and humans and have been linked with MPs. This species has been proven to initiate biofilm and promote the further attachment of other bacterial species.53 Recently, pathogenic Vibrio parahaemolyticus has been isolated from MP particles such as polyethylene, polypropylene and polystyrene from North/Baltic Sea. In addition, V. cholerae, and V. vulnificus also been found to reside on the surface of polypropylene (PP) and polyethylene (PE) MPs.54 These MPs are poorly degradable and enable the bacteria to colonize rapidly. Compared to natural plant or animal-based polymers, these are easily transported over long distances, carrying microbes.54
A fish pathogen, Aeromonas salmonicida and Campylobacter spp. has been detected in DNA isolated from MPs obtained from the North Adriatic sea by high throughput and metagenomic techniques.55 PE MPs exhibits hydrophobic characteristics in water and can agglomerate pathogens rather than disperse in the aqueous medium. PE MPs with high concentrations of E. coli were shown to adsorb and aggregate on the surface within 30 seconds. DNA samples from in situ tests of PE MP beads in river Barrow, Carlow, Ireland revealed that Enterobacter spp., Helicobacter spp., Arcobacter spp., Clostridium perfringens, and E. coli were present.56 It shows MPs are prone to attract enteric pathogens like E. coli and other pathogens in the aqueous medium and resist inactivation by disinfectant. Hence, agglomerated enteric pathogens on PE may pose a serious public health impact.57 However, different pathogens showed diverse preferences for adsorption on microplastic substrates. This was highly influenced by temperature and the availability of inorganic salt like nitrate. Polyethylene terephthalate (PET) was most favored by pathogens such as Vibrio, Tenacibaculum, and E. coli than polyethylene (PE), and polypropylene (PP) in an aqueous medium.58 Enteric viruses are available in large quantities on the surface water of the environment. The association of these viruses with plastic spheres could potentially cause infections in humans. Recently, the enveloped bacteriophage (Phi6) and enteric virus-like rotavirus (RV) SA11 have been tested in different water, such as sterile, environmental, and with added nutrients. Viruses are strongly associated with biofilm-colonized pellets. It protects itself from getting inactivated and stable for more than two days. Hence, viruses that colonized the MPs may have a potential public health risk of water when coming in contact with a susceptible host.59
There were thirteen gastrointestinal parasite taxa present in three wild populations of capybaras in the west-side lowlands of the Orinoco Basin. These included five protozoans of the phyla Apicomplexa, Amoebozoa, and Ciliophora, and eight metazoans of the phyla Nematoda and Platyhelminthes. However, no significant relationship was reported between parasites and MPs.60 Recently, it has been demonstrated that enteric parasites such as Toxoplasma gondii, Cryptosporidium parvum, and Giardia enterica could adhere to microfiber surfaces compared with microbeads. These pathogens are persistent in the marine environment, and the association of these with MPs mediates the transmission of the parasite from wildlife to humans.61
Research suggests that the gut microbiome is responsible for good human health and lifestyle-related diseases.62,63 Commensals of gastrointestinal microorganisms can potentially secrete lipolytic esterases such as lipases, esterases, cutinases, and PETases, which convert nutrients into end products. Recently, Lactobacillus rhamnosus GG (ATCC 53103) and Bifidobacterium longum (NCC 2705) has shown to secrete the acyl hydrolase/lipase and to cleave esters of fatty acids into their metabolites.64 Bacterial metabolizing enzymes are versatile and work differently under diverse conditions. Bacteria can use metabolizing enzymes as biocatalysts to degrade microplastics, particularly esters of microplastic residues.65 Recently, Acinetobacter baumannii, Bacillus weihenstephanensis, Pseudomonas aeruginosa, Pseudomonas fluorescens, and Rhodococcus ruber have been shown to exhibit the breakdown of microplastics into microbial biomass and gases.65 Bacteria evade the action of microplastics by secreting inherited metabolizing enzymes like hydrolases and lipases. Some bacteria secrete lipases that allow them to survive in various niches, escape the host's defenses, thermotolerant, hydrophobic and metabolize xenobiotics such as esters of microplastic residues.65–67 In opportunistic human diarrheal pathogens, lipases are considered essential virulence traits. Pathogenic Vibrio species adapt to various niches for survival, including host cells and a protective environment by secreting metabolizing enzymes of esterases/lipases.67
MPs in the dissemination of antibiotic resistance
MPs solubilized in the water are effective carriers of the environmental microbes and antimicrobial resistance bacteria (ARB), making them likely to be continuously imported into the aquaculture environments and by humans. The aquatic environment serves as an excellent reservoir for ARG, heavy metals, and dissolved organic substances that promote bacteria to form biofilm. MPs particles provide a hydrophobic nature in the environment that facilitates the adsorption of bacteria and selectively enable the bacteria to form biofilm. In addition, MPs can absorb pharmaceuticals, including antibiotics, biocides, and heavy metals. Absorption of these agents on the MPs containing biofilm may lead to tolerance of selective pressure and the emergence of ARB. Low concentrations of antibiotics, biocides, and heavy metals can help bacteria become antibiotic resistance phenotypes and induce horizontal ARG transfer.68 Recent research has demonstrated that many of these chemicals, such as metallic nanoparticles, non-antibiotic drugs, and non-antibiotic conditions of MPs could accelerate the dissemination of ARGs and are highly favorable for the evolution of ARGs.69 Bacterial conjugative transfers can be accelerated by up to 1.7 folds by microplastics, which aggravates ARG spread. Additionally, MPs regulates genes encoding outer membrane proteins and plasmids needed for conjugation.70 Moreover, horizontal gene transfer among the bacteria is more efficient in the biofilm form of bacteria in biofilms than planktonic bacteria.71Transfer of antibiotic resistance genes between microorganisms on the surface of plastics may favor the persistence of pathogens in the environment. The natural aging of plastics in the soil leads to plastic vibration, fragmentation, and translocation. MPs aged by UV light, temperature, and chemicals increase bacterial adsorption and ARG transfer synergistically. Micro and nano-sized rod-shaped plastic fibers could travel downward from the surface of loamy soil. The antibiotics tetracycline, beta-lactam and sulphonamide were detected in the plastic recover from penetrated soil due to hetero-aggregation of soil minerals with MPs and antibiotics were identified as a driving mechanism for MPs translocation.72 Biodegradable MPs broadened the host range for antimicrobial resistance genes and propagated resistance genes among diverse bacterial populations such as Proteobacteria, Bradyrhizobium, and Pseudomonas.73 MPs present in the freshwater, coastal, and deep ocean sediments were selectively enriched genus Afipia and other bacteria of Nitrobacteraceae to colonize MPs. These nitrifiers, denitrifiers, and ammonia oxidizer significantly altered the concentration of NO3−, NO2−, and NH4+ in MPs. These chemical alterations could be directly related to promoting the efflux pump system on bacteria. The reactive oxygen species generated may be mediated by DNA damage, increased efflux, and stress response induced by MPs exposure could promote ARG transfer.74,75 MPs like polyethylene terephthalate increase bacteria's resistance to antibiotics, such as Vibrio, Muricauda, and Ruegeria. These organisms showed resistance to penicillin, sulfafurazole, erythromycin, and tetracycline at high levels of MPs. Several gene cassettes are associated with class 1 integrons in bacteria that contain MPs compared to bacteria that do not.76 Sludge anaerobic digestion increases the antimicrobial resistance genes. It promotes the spread of antimicrobial resistance genes through mobile gene genetic elements at a concentration of 10 mg L−1, which is a hundredfold higher environmental concentration of dimethyl phthalate in the water.77 In sewage sludge thermophilic anaerobic digestion (AD), PE-MPs selectively enriched acetogens Thermoanaerobacter and hydrolytic bacteria Caldicoprobacter, which may be hosts for ARGs and MGEs.78 Recently, UV-aged polystyrene MPs (PS-MPs) have been demonstrated to understand the aging process of MPs and ARG transfer efficiency. After twenty days of UV exposure, PS-MPs exhibited 5-8-fold increased adsorption capacity for E. coli carrying ampicillin, carbapenemase, and streptomycin resistance plasmids compared to pristine PS-MPs. UV exposure to PS-MPs can depolymerize PS-MPs, which could induce intracellular reactive oxygen species generation, increase cell permeability and up-regulate horizontal gene transfer associated genes.79 Polyvinyl chloride (PVC), and polyamide (PA) can stimulate and support extracellular polymeric substances and reactive oxygen species. In aerobic granular sludge systems, these MPs increase intracellular and extracellular ARGs such as the tetracycline resistance genes tetW and tetE and class 1 integrase genes (intI1) in potential pathogenic bacteria.80 It has been demonstrated that class 1 integron integrase genes (intI1) and bacterial communities associated with microplastic biofilms under antibiotic sulfamethoxazole (SMX) pressure increased relative abundance of ARGs on the two different MPs. SMX stress increased the abundance of mobile genetic elements like integrons at high concentrations and promoted the dissemination of ARGs at low concentrations after prolonged incubation.81
Mitigation of MPs in the environment by bioremediation
Biodegradation of phthalates
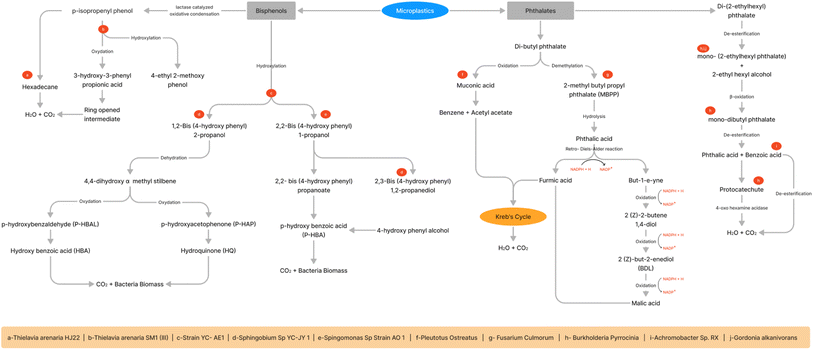
Several microorganisms isolated from diverse sources (Table 1) such as wetland soil, contaminated soil pesticides, recycled paper industry, water, sediments, sewage treatment plant, tap water of the public toilet, and marine plastic debris tested for degradation potential of MPs persists in the environment.82 The major pathways of bioremediation of phthalates and biophenols are shown Fig. 4. Different substrates were used inorganic salts, glucose, peptone, beef extract, yeast extract, tryptone, and beef extract plus peptone at low and high concentrations. Among these substrates, combination of beef extract and peptone had the highest removal efficiency. The pathways were identified as β-scission, debromination, and nitration routes.83Arthrobacter sp. isolated from wet soil was tested on di-n-butyl phthalate (DBP) as a carbon source and converted to phthalic acid. However, alkyl chain length, molecular weight, and concentration influenced biodegradation.84 An environmental saprophytic bacterium, Agromyces spp. utilized a phthalate (DEHP) as a carbon source at room temperature (29.6 °C) with a pH of 7.2. This bacterium converted mono-ethylhexyl phthalate into phthalic acid after incubation for a week with a substrate density of 200 mg L−1.85Ochrobactrum anthropi bacteria at a similar concentration of phthalate (DEHP) can break down 98.7% of DEHP within 72 h.86Gordonia alkanivorans utilized DEHP as the sole carbon. This bacterium can sustain high DEHP concentrations (from 100 to 800 mg L−1), and the degradation rates were all above 94% when the temperature was at 30 °C and pH 8.0. It also acts as a halotolerant and can withstand up to 0–5% NaCl in trace element media augmented with DEHP, while the DEHP breakdown rates reduced with increasing NaCl content.87 Halotolerant bacterial consortia contained Gordonia sp. (54.93%), Rhodococcus sp. (9.92%), and Achromobacter sp. (8.47%) degraded di-(2-ethylhexyl) phthalate (DEHP) in medium enriched from activated sludge. It could degrade 93.84% of 1000 mg L−1 DEHP after 48 h incubation with the intermediates, including (2-ethylhexyl) phthalate, mono-ethylhexyl phthalate, mono-hexyl phthalate, and mono-butyl phthalate.88 Other microbial consortia consist of Rhodococcus, Niabella, Sphingopyxis, Achromobacter, Tahibacter, and Xenophilus obtained from farmland soil degraded DEHP concentration of 1000 mg L−1 within 24–72 h by de-esterification and β-oxidation mechanism.89Achromobacter sp. isolated from activated sludge degraded DEHP with Tween-80 as a solubilizing agent. Masson pine seed powder was used as a co-metabolic substrate to increase biodegradability at 30 °C and pH 7.0. The biodegradation of DEHP began with mono-(2-ethylhexyl) phthalate (MEHP), which was then further de-esterified to produce phthalic acid and benzoic acid. In the end, benzoic acid mineralized to CO2 and water.90Burkholderia pyrrocinia can degrade 98.05% of 500 mg L−1 of DEHP in the presence of 1% yeast extract in a mineral salt medium with pH 7.0 and temperature of 30 °C. Using yeast extract as an additional nutrient and mineral source significantly increased the bacterial growth, concentration, and surface area, leading to a higher DEHP biodegradation ratio. The enzyme esterase secreted from bacteria first break down DEHP to MEHP, which is subsequently transformed to MBP by β-oxidation. Then, esterase breaks down MBP to PA, quickly transforming into protocatechuate (PCA) in an aerobic environment. PCA is ultimately cleaved to produce 4-oxo-hexanoic acid, CO2, and H2O.91Rhodovulum sp. degraded di-butyl phthalate (DBP) of concentration at 600 mg L−1 into diethyl phthalate (DEP), and mono-n-butyl phthalate (MBP) was the intermediate compounds. It utilized sugar substrate as a major nutrient and showed enhanced growth with a degradation rate of above 75% with a concentration of 5.0 g L−1. The carbon source influenced the degradation rate (glucose > fructose > sucrose > maltose > lactose > citric acid > starch).92Pleurotus ostreatus could metabolize DBP at a high concentration of 1000 mg L−1 after 504 h into acetyl acetate and benzene, the latter of which would enter the Krebs cycle after being oxidized to muconic acid.93Pseudomonas sp. degrade DBP into phthalic acid, salicylic acid, 3-hydroxybenzoate acid, 3-carboxy-cis-cis-muconate, and fumarypyravate.94Karenia brevis can degrade phthalates like DMP, DEP, DAP, and DPrP by de-esterification, demethylation or transesterification pathways and their corresponding metabolites were dimethyl phthalate (DMP), monoethyl phthalate (MEP), mono-methyl phthalate (MMP), phthalic acid (PA), protocatechuic acid (PrA) and diethyl phthalate (DEP).95 A fungus Fusarium culmorum isolated from the recycled paper industry, degraded DBP at a concentration of 1000 mg L−1 after incubation of 228 h by demethylation to yield 2-methylbutylpropyl phthalate (MBPP) and phthalic acid (PA). Then, by a reverse Diels–Alder reaction and the electron resonance effect, PA would be metabolized into two separate substances: fumaric acid (FA) and but-1-e-yne (BE). FA can go through the Krebs cycle and break down into CO2 and water. BE would oxidize into (2Z)-2-butene-1,4-diol (BD). BD would oxidize to (2Z)-but-2-enedial (BDL). BDL undergoes oxidation to produce malic acid (MA).96 A Comamonas testosteroni can degrade up to 1000 mg L−1 of para-toluic acid after 14 h into terephthalic acid and phthalic acid. The addition of yeast extract as a nitrogen source enhanced the degradation reaction.97Diaphorobacter sp., a Gram-negative bacterium, utilize phenanthrene (PHE) as sole carbon and degrade 96.3% of 100 mg L−1. It can withstand maximum of 40 °C and 400 mg L−1. While the addition of salicylic acid and 2-hydroxynaphthalene could hinder the growth of this organism, the addition of phthalic acid enhanced the strain's ability to degrade. The degradation mechanism through ring cleavage yields a product of 1,2-dihydro-acenaphthylene and enters the phthalic acid pathway. However, 1,2-dihydro-acenaphthylene breaks down immediately to 2-isopropyl-5-methylphenol, which subsequently transforms into β-resorcylic acid in the TCA cycle.98Rhizobium petrolearium can degrade a polynuclear hydrocarbon phenanthrene via two pathways of naphthalene and the phthalic acid routes. This bacterium had a specific gene encoding for the degradation of aromatic compounds through the meta pathway. Initially, deoxygenation was on 3,4-carbon and followed by degradation by meta cleavage yield 1-hydroxy-2-naphthoic acid. Then, it is broken down by the phthalic acid and naphthalene pathways.99 Polycyclic aromatic hydrocarbons (PAHs) were degraded by soil microbial consortium (OMC). It was able to degrade naphthalene, acenaphthylene, acenaphthene, fluorene, and phenanthrene in solution to 100%, as well as anthracene and fluoranthene to up to 76% and 50%, respectively. A novel Stenotrophomonas maltophilia CPHE1 identified to mineralize a polycyclic Aromatic Hydrocarbons into salicylic and phthalic acid.100| S. No. | Microorganism | Source | Microplastic | Method/conditions | Biocatalyst/% of degradation | Metabolites/end point | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Paraconiothyrium variabile | Archived culture, Iran | Phenol and bisphenol A | pH: 5; temperature (temp): 50 °C; duration: 30 min | Laccase/96–88 | Biomass | 106 |
| 2 | Chlorophyta picocystis | Household sewage, Centre East of Tunisia | Bisphenol A | Light/bioremediation temp: 37 °C; light intensity: 80.60 mmol photons m−2 s−1; inoculums size: 25 × 105 cells per ml; duration: 120 h | Photolytic and microalgae/91 | Biomass | 114 |
| 3 | Aspergillus terreus and Aspergillus flavus | The gas station, paint industries, pesticides wastes in Egypt | Bisphenol A | Temp: 30 °C; pH: 5–7; duration: 6 days | Cellular metabolism/40–50 | Thiopropionamide, methanone; 3-amino-2-benzofuryl)(4-chlorophenyle); 1H-pyrazole; 4,5-dihydro-5,5-dimrthyle-4-isoprpylidene, phenol; 2,4-isopropylidenedi; phenol; 2,6-bis(1,1-dimethylethyle)-4-(methyle-1-phenylethyle), bis (2-ethylehexyle)phthalate | 108 |
| 4 | Sphingobium sp. | River sludge, Guangdong Province, China | Bisphenol A 50–300 mg L−1 | Temp: 30 °C; pH: 6.5; duration: 5 days | CYP monooxygenases/90 | 1,2-Bis(4-hydroxyphenyl)-2-propanol, 4,4′-dihydroxy alpha methylstilbene, 4-hydroxybenzaldehyde, 4′-hydroxyacetophenone, 2,3-bis(4-hydrophenyl)-1,2-propanediol | 115 |
| 5 | Chlamydomonas reinhardtii | Culture collection from National Learning Service, Colombia | Bisphenol A 25–100 mg L−1 | Temp: 24 ± 2 °C; fluorescent lights: 5000 lux; duration: 12 h of light and 12 of darkness; aeration: 0.7 vvm using atmospheric air | Cellular metabolism/96–98 | Biomass | 116 |
| 6 | Sphingomonas bisphenolicum | Soil, Yawata city, Kyoto Prefecture | Bisphenol A | Temp: 30 °C; duration: 7–35 days | Cellular metabolism | p-Hydroquinone; 4-hydroxyacetophenone; 4-hydroxybenzoic; 4-isopropenylphenol | 107 |
| 7 | Lactococcus lactis, Bacillus subtilis, Lactobacillus plantarum, Enterococcus faecalis, and Saccharomyces cerevisiae | S. cerevisiae from Cell Biology Research Center, Montpellier, France; Lactococcus sp., Bacillus sp. and Enterococcus sp. from Aristotle University of Thessaloniki, Greece | Bisphenol A 2500 μg | Temp: 30 °C; duration-96 h | Minimal essential medium with BPA as carbon source/40–50 | Hydroquinone (HQ); 4-hydroxyacetophenone (HAP); 4, hydroxybenzoic acid (HBA); 4-iso-propenylphenol (PP) | 45 |
| 8 | Chaetomium strumarium, and Thielavia arenaria | Soil, South Tunisia | Bisphenol A | pH: 6.0; temp: 35 °C duration: 4 days | Laccase and dye decolorizing peroxidase | Biomass | 117 |
| 9 | Shewanella haliotis | Soil sediment form estuarine system, Brazil | Bisphenol A 150 mg L−1 | pH: 7.4; temp: 30 °C; duration: 10 h | Bisphenol-hydroxylase-A | 4-Hydroxyphenyl alcohol, 2,2-bis(4-hydroxyphenyl)-1-propanol, 2,3-bis(4-hydroxyphenyl)-1,2-propanediol | 118 |
| 10 | Pseudomonas putida | Soil samples from Heilongjiang, Hebei, Shandong, Guangdong and Anhui, China | Bisphenol A 0.5–1000 mg L−1 | pH: 7.2; temp: 30 °C; duration: 72 h | Cellular metabolism | 4,4-Dihydroxy-alpha-methylstilbene; p-hydroxybenzaldehyde; p-hydroxyacetophenone, 4-hydroxyphenylacetate; 4-hydroxyphenacyl alcohol; 2,2-bis(4-hydroxyphenyl)-1-propanol; 1,2-bis(4-hydroxyphenyl)-2-propanol; 2,2-bis(4-hydroxyphenyl) propanoate | 109 |
| 11 | Enterobacter gergoviae | Khuzestan petrochemical wastewater, Iran | Bisphenol A 45 mg L−1 | pH: 7.0; temp: 30 °C; duration: 12 days | Cellular metabolism | Biomass | 119 |
| 12 | Trametes versicolor | Rotten Wood, Zhejiang University, China | Bisphenol A | pH: 4.0–8.0; temp: 30 °C; duration: 7 days | Laccase/90 | Biomass | 120 |
| 13 | Escherichia coli | Bioengineered E. coli | Diisobutyl phthalate 1.5 mg mL−1 | Citrate/phosphate buffer (pH 5.0–8.0), Tris/HCl (pH 8.0–9.0) and boric acid/borax (pH 9.0–10.0); temp: up to 80 °C; duration: 10 days | The whole cell biocatalyst producing carboxylesterase | Biomass | 111 |
| 14 | Agromyces sp. | Soil, Guangzhou, China | Di-(2-ethylhexyl) phthalate 50–1000 mg L−1 | pH: 7.2; temp: 29.6 °C; inoculum size: OD600 of 0.2; duration: 12 days | The whole cell biocatalyst | Mono-ethylhexyl phthalate, phthalate acid | 85 |
| 15 | Arthrobacter sp. | Soil, and water, Liaoning Province, China | Phthalates 10–300 mg L−1 | pH: 7.0; temp: 30 °C; duration: 7 days | DBP-10 mg L−1-32 h, 300 mg L−1-25 h, DMP-99.5, DEHP-51.4, PA-81 | Biomass | 84 |
| 16 | Ochrobactrum anthropi | Water and soil from Wuhan South Lake, Wuhan, China | Di-2-ethylhexyl phthalate 400 mg L−1 | pH: 6.0; temp: 30 °C; NaCl: 10 g L−1 | The whole cell biocatalyst/98 | Biomass | 86 |
| 17 | Karenia brevis | Archived culture from Institute of Oceanography, Chinese Academy of Sciences, China | Dimethyl phthalate (DMP), Diethyl phthalate (DEP), Diallyl phthalate (DAP), Dipropyl phthalate (DPrP) concentration, DMP: 0.257 mmol L−1, DEP: 0.135 mmol L−1, DAP: 0.102 mmol L−1, DPrP: 0.060 mmol L−1 | Culture incubated in light/dark cycle of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 h at 23 ± 1 °C with 4000 lx illumination in an intelligent artificial climate 10 h at 23 ± 1 °C with 4000 lx illumination in an intelligent artificial climate |
Esterases, dialkyl hydrolase DMP-87.1, DEP-61, DAP-46, DPrP-40 | Dimethyl phthalate, monoethyl phthalate, phthalic acid | 95 |
| 18 | Gordonia alkanivorans | Soil, Heze city, Shandong Province, China | Di(2-ethylhexyl) phthalate 100–800 mg L−1 | Temp: 30 °C; pH: 7.4; NaCl: 0–5% | The whole cell biocatalyst/94 | Phthalic acid, mono (2-ethylhexyl) phthalate, benzoic acid | 87 |
| 19 | Fusarium culmorum | Culture Collection, Research Centre for Biological Sciences, Universidad Autonoma de Tlaxcala, Mexico | Dibutyl phthalate 500–1000 mg L−1 | pH: 7.4; temp: 28 °C; duration: 10 days | The whole cell biocatalyst/99 | Fumaric and malic acid, but-1-e-yne, CO2 and H2O | 93 |
| 20 | Burkholderia pyrrocinia | Soil, China | Di(2-ethylhexyl) phthalate 500 mg L−1 | pH: 2–7.0; temp: 30 °C; duration: 6 days | The whole cell biocatalyst/95 | Mono(2-ethylhexyl) phthalate, mono-dibutyl phthalate, phthalic acid, 4-oxo-hexanoic acid, CO2 and H2O | 91 |
| 21 | Achromobacter sp. | Activated sludge, Nanjing Jiangxinzhou Sewage Treatment Plant, China | Di(2-ethylhexyl)phthalate | pH: 7.0; temp: 30 °C; duration: 96 h | The whole cell biocatalyst/86–91 | Mono(2-ethylhexyl) phthalate, phthalic acid, benzoic acid, CO2 and H2 | 90 |
| 22 | Arthrobacte. globiformis, Pseudomonas putida, Pseudomonas hunanensis | Soil, Fragrant Hills Park of Beijing, China | Phthalic acid | pH: 7.4; temp: 28 °C; duration: 36 h/dark | The whole cell biocatalyst/99 | Biomass | 121 |
| 23 | Bacterial consortium Pandoraea sp. and Microbacterium sp. | Activated sludge, Guangzhou city, China | Dibutyl phthalate 35–500 mg L−1 | pH: 5.5–8.5; temp: 30 °C; duration: 72 h | The whole cell biocatalyst/>90 | Dibutyl phthalate, phthalic acid | 122 |
| 24 | Rhodovulum sp. | Water, Jeddah, Saudi Arabia | Di-n-butyl phthalate 200–1000 mg L−1 | pH: 5.0–9.0; duration: 1–5 days; NaCl: 1–5%; glucose: 5.0 g L−1 | The whole cell biocatalyst/>90 | Diethyl phthalate, mono-n-butyl phthalate | 92 |
| 25 | Bacillus marisflavi | Tap water, Guru Nanak Dev University, Amritsar, India | Benzyl butyl phthalate, dimethyl phthalate | pH: 7.0; temp: 35 °C; NaCl: 1% | Esterases/hydrolase; permeases; phthalate 4,5-dioxygenase; cis-phthalate; dihydrodiol dehydrogenase; 4,5-dihydroxyphthalate decarboxylase; 3,4-dihydroxygenase; 3,4-dihydroxyphthalate decarboxylase; protocatechuate 4,5-dioxygenase | Monobutyl phthalate, monobenzyl phthalate, phthalic acid, carbon dioxide, water | 82 |
| 26 | Comamonas testosteroni | Soil from agricultural fields, Chongqing, China | Dimethyl phthalate 450 mg L−1 | pH: 9.0; temp: 30–35 °C; duration: 14 h | 2-Pyrone-4,6-dicarboxylic acid hydrolase (LigI); 99 | Mono-methyl phthalate, phthalic acid | 123 |
| 27 | Gordonia terrae | Soil, Guangdong Ocean University | Di-(2-ethylhexyl) phthalate | pH: 5.0; temp: 30 °C; duration: 72 h | Hydrolase/91.8 | Phthalic acid, mono (2-ethylhexyl) phthalate, 2-ethylhexanol (2-EH) | 50 |
| 28 | Pseudomonas sp. | Soil, Harbin, China | Dibutyl phthalate 100–2000 mg L−1 | pH: 7.0; temp: 35 °C; duration: 48 h | The whole cell biocatalyst/90 | Phthalic acid, salicylic acid, 3-hydroxybenzoate acid, 3-carboxy-cis,cis-muconate, fumarypyravate | 33 |
| 29 | CM9 consortium Rhodococcus, Niabella, Sphingopyxis, Achromobacter, Tahibacter, Xenophilus | Soil, suburban farmland in Shanghai, China | Di-(2-ethylhexyl) phthalate 1000 mg L−1 | pH: 7.4; temp: 30 °C; duration: 24–72 h | Whole cell biocatalyst/90–100 | Mono(2-ethylhexyl) phthalate, diethyl phthalate, dimethyl phthalate, phthalic acid, phthalic acid (2-ethyl hexyl-methyl ester), phthalic acid (2-ethyl hexyl-hexyl ester | 89 |
| 30 | Pleurotus ostreatus | Culture collection, College of Postgraduates, COLPOS Puebla, Mexico | Dibutyl phthalate 500–1000 mg L−1 | pH: 7.4; temp: 21 °C; duration: 21 days | Esterase, decarboxylase, NADH dehydrogenase, oxidase, alcohol dehydrogenase, aldehyde dehydrogenase, dehydratase/94–99 | Benzene, acetyl acetate, muconic acid | 96 |
| 31 | Burkholderia cepacia, Pseudomonas koreensis, Ralstonia pickettii | Santos Estuarine System, Brazil | Diethyl phthalate 300 mg L−1 | pH: 7.0; temp: 30 °C; NaCl: 2.5%; duration: 24 h | Esterases, permeases, dehydrogenases, dioxygenases | Biomass | 124 |
| 32 | Cyanobacteria, Synechocystis sp. PCC6803, Synechococcus sp. and Cyanothece sp. | Culture collection, Purdue University, West Lafayette, USA | Dimethyl phthalate 20–50 mg L−1 | pH: 9.0; temp: 30 °C; duration: 120 h | Esterases | Phthalic acid | 125 |
| 33 | Halotolerant bacterial consortium | Activated sludge, Xin Xiang, Henan Province, China | Di-(2-ethylhexyl) phthalate 100–2000 mg L−1 | pH: 6.0, temp: 30 °C; NaCl: 0–3%; duration: 72 h | The whole cell biocatalyst/92 | 2-Ethylhexyl pentyl, butyl(2-ethylhexyl phthalate), mono ethylhexyl phthalate, mono-hexyl phthalate, monobutyl phthalate | 88 |
| 34 | Sphingobium yanoikuyae | Soil, Jianghan plain, Hubei, China | Phthalate esters 8.5–50 mg L−1 | pH: 7.5; temp: 30 °C; duration: 10 days | Phthalate dioxygenase | Biomass | 126 |
A Basidioascus persicus metabolized pyrene, phenanthrene and anthracene in the presence of 2.5% minimal medium and in the presence of Persian Gulf Medium the removal rate increased to 19-fold. Mono- or dioxygenases enzymes produced by bacteria can convert pyrene to hydroxypyrene by forming a hydrodiol. Then, in the subsequent transformation of hydroxypyrene into hydroxy naphthoic acid by distinct cleavage pathways. Hydroxy naphthoic acid broke down into phthalic acid, protocatechuic acid, and salicylic acid by tricarboxylic acid cycle.101 A Comamonas sp. degraded tetrabromobisphenol A (TBBPA) at a concentration of 50 mg L−1 within ten days of incubation. The dehalogenation of TBBPA yields BPA, 3,4-dihydroxymandelic acid, beta resorcylic acid, acetylbenzoyl, acetophenone, and oxalo-acetic acid metabolites were reported.102 Another metabolic products were 2,6-dibromo-4-(1-methyl-1-phenylethyl) phenol, 2-bromophenol by debromination, and phenol in the biodegradation of TBBPA.103Phanerochaete chrysosporium removed TBBPA within 12 h by glycosylation and oxidative hydroxylation. The fungal transformation identified TBBPA metabolites such as tetrabromobisphenol A glycoside, tribromobisphenol A glycoside, and monohydroxylated tetrabromobisphenol A.104 A marine consortium containing Limnobacter, Pseudoalteromonas, Glaciecola, Thalassomonas, and Alteromonas degraded the TBBPA in 10 days under aerobic metabolic conditions, and efficiency was 90% degradation at a concentration of 10 mg L−1. Irpex lacteus yield four metabolic substrates: phenol, glucose, sodium pyruvate, and sodium citrate. This organism can biodegrade TBBPA at a wide pH range of 4.0–12, and maximum degradation occurred at a pH of 12.00 with sugar-like glucose and sodium pyruvate enhanced. Seven intermediates found during biodegradation of TBBPA such as 1-(3-bromo-4-hydroxyphenyl)ethanone, 4-hydroxybenzoic acid, bisphenol A, 2,5-dihydroxyacetophenone, 2,5-dihydroxybenzoic acid, 2,4-dihydroxyacetophenone, and 2,4-dihydroxybenzoic acid.105 The enzyme laccase purified from Paraconiothyrium variabile (PvL) removed 80% of phenol and 59.7% of bisphenol A at temperature of 50 °C and pH 5.0.106
Biodegradation of bisphenols
Bisphenol A (BPA, 2,2′-bis(4-hydroxyphenyl)propane) was degraded by microbial consortium of Sphingomonadales, Xanthomonadales, Burkholderiales and Pseudomonadales isolated from soil.107Saccharomyces cerevisiae, Lactococcus lactis, Bacillus subtilis, Lactobacillus plantarum, and Enterococcus faecalis are the five possibly probiotic microorganisms that can eliminate BPA. The degradation resulted in the formation of two dimer products in cells while the compounds Hydroquinone (HQ), 4-Hydroxyacetophenone (HAP), 4-Hydroxybenzoic acid (HBA) and 4-iso-propenylphenol (PP).45 Fungal isolates such as Aspergillus terreus and A. flavus degraded BPA with sodium nitrate at pH 5.0 and yeast extract at pH 7.0 respectively and incubation temp at 30 °C after incubation at six days at shaking state. The following were the intermediate products of BPA thiopropionamide, methanone, (3-amino-2-benzofuryle)(4-chlorophenyle), 1H-pyrazole, 4,5-dihydro-5,5-dimrthyle-4-isoprpylidene, phenol, 2,4-isopropylidenedi, phenol, 2,6-bis(1,1-dimethylethyle)-4-(1-methyle-1-phenylethyle), bis(2-ethylehexyle) phthalate.108Pseudomonas putida isolated from soil had high-level biodegradation of hydrocarbons within 72 hours. The metabolites identified such as follows 4,4-dihydroxy-alpha-methylstilbene, p-hydroxybenzaldehyde, p-hydroxyacetophenone, 4-hydroxyphenylacetate, 4-hydroxyphenacyl alcohol, 2,2-bis(4-hydroxyphenyl)-1-propanol, 1,2-bis(4-hydroxyphenyl)-2-propanol and 2,2-bis(4-hydroxyphenyl)propanoate.109Enzymatic degradation of microplastics
Biological degradation of microplastic in microbes (bacteria and fungi) can occur through aerobic and anaerobic mechanisms. Microbes can convert microplastic into biomass like CO2, water, and in some cases, lower member of hydrocarbon like methane. The majority of biodegradation processes are designed to work in aerobic conditions. It is best suited to degrade microplastic in different environmental conditions. However, anaerobic degradation requires controlled conditions and specialized equipment. Aerobic biodegradation of microplastics may be carried out with a single microbe or consortium of microbes. These microbes may release extracellular enzymes like carboxylesterase, hydrolase, lipases, catalases, and oxygenase enzymes, which are functional group-specific cleavage of microplastics that lead to the conversion of polymeric forms into the monomeric form of microplastic metabolites.110 The release of these enzymes from wild-type microbes is significantly fewer, making this process slow. Recently, carboxylesterase has surfaced anchored on the non-pathogenic cell wall and demonstrated as the whole cell for degradation of microplastics.111 Anaerobic degradation usually takes in the intracellular milieu. Microplastic may penetrate cells through the efflux or diffusion mechanism, metabolized by intracellular enzymes through oxidation and the tricarboxylic acid (TCA) cycle, and finally be mineralized into oxidised products like carbon dioxide, water and hydrocarbon like methane.110 The consortium of biodegradation microbes may offer various intracellular and extracellular enzymes to degrade microplastics. However, very few enzymes have been identified for the degradation of microplastics.112 Sometimes, aggregation of microbes in the presence of microbes leads to ageing and loss of the crystalline structure of microplastic.Addition of enzymes into the biodegradation process with microbes may enhance the degradation process and wide range of microplastic residues. The blend of bioremediation with microbes and enzyme technology is the preferred method for completely removing microplastics within a short duration in wastewater treatment. However, it has been demonstrated in bench scale, and real practical applications are not in practice.112 This could be due to microbes' limited availability with region-specific and variable efficiency in degrading microplastics. Enzymes are less stable, narrow specific, and offer regio-selective degradation. To overcome these challenges, entrapping enzymes at metal–organic frameworks offer better activity, stability, and recycle potentials in degrading organic micropollutants in the environment.113 Again, it has to be transformed from a lab scale into industrial applications.
Advantages of bioremediation
Microorganisms like algae, bacteria, and fungi were explored to remove the xenobiotics from the field in purified enzymes or immobilized form or biosorption of organic pollutants methods.106 Among the microorganisms, fungi were the most effective at detoxicating xenobiotics in the environment due to their adaptability and ability to tolerate diverse conditions. Microorganisms are often used for biodegradation under extreme conditions and high anthropogenic pollution.114 In addition, a consortium of microorganisms is used for the highest level of biodegradation. Microorganisms use aerobic, anoxic and anaerobic methods for the remediation of xenobiotics by either oxidation or reduction reactions. Secretory enzymes like carboxylesterases or hydrolases accept a wide range of substrates for degradation with selectivity and specificity in degradation and provide stability in the presence of organic solvents.127 The use of a consortium of bacteria in bioremediation increases biodegradation efficiency in a variety of ways. The synergistic effect of multiple biochemical pathways and oxidoreductases offers the most incredible efficiency in degradation.128 Ultimately, bioremediation results in either bio-transformed or mineralized biomass, which is a green method for removing xenobiotics. Researchers use a combination of metabolomic and genomic approaches to elucidate the probable metabolism of specific toxicants during degradation. It will enable identifying potential modes for bioengineering desired microorganisms for effective degradation.Challenges associated with bioremediation
All the bacteria might not have the potential to metabolize a broader range of microplastics. There are significant difficulties in removing low-level microplastics from the environment due to the low-level expression of genes, and at high concentrations, bacterial growth may be inhibited.86 Although the biodegradation of microplastics was extensively studied, the knowledge of metabolic mechanisms, biocatalysts, and expression of genes is still limited. Expression of non-specific genes in fungi-based bioremediation of microplastics affects the efficiency. The degradation efficiency of microplastics is affected by several factors like temperature, pH, inoculum size, and NaCl concentration which is challenging to maintain in large-scale degradation.127 The large-scale practical application of bioremediation is complex due to the purification costs, low catalytic activities, and poor enzyme stability of the metabolizing enzymes.127 The complex and transient action of microbial metabolic processes makes the detection of intermediate metabolites intricate. There is the possibility of misdetection of some metabolites, and non-relevant metabolic reactions could occur. End products of some oxidation reactions lead to dihydro diol epoxides, which may be bioactive and toxic. There is an urgent need for more experimental ideas for subtle, quick detection of reaction end products through advanced methods.Conclusion
In recent years, the quantum of research work on MPs has increased many folds. Human exposure to plastics and their secondary metabolites, like micro and non-sized plastics, is inevitable. There is growing scientific evidence for MPs in human blood, lungs, and the placenta. MPs in human organs are evidenced by fluctuating metabolic activities, growth, reproduction, and cancer induction. Still, there are potential research avenues on the health impacts of toxic chemical additives and pollution in plastics that threaten the environment and human health worldwide. The environment setting remains a source and reservoir for MPs and plastic additives. However, the biotransformation and chemical transition of MPs are mainly unknown. Absorption, desorb, and physicochemical characteristics of native and transformed MP are not known and require hyphenated techniques to detect them. MPs present in the environment will have a potential impact on human health. Everyone eats and inhales some quantum of MPs, and there is a lack of knowledge on the pharmacokinetics of MPs in humans. MPs are immortal in the laboratory were remediated by region-specific or laboratory-specific microbes. We found that photocatalysts can degrade persistent organic pollutants, pharmaceuticals and enable disinfection by generating radicals that can break highly energetic chemical bonds with low energy input, especially when solar radiation is used.129–133 Thus, they present themselves as a potential technology for the degradation of plastic and the toxicity associated with their presence in water in remote areas. The use of advanced oxidation processes (AOP) is growing, especially when addressing the recalcitrant contaminants and pollutant degradation. While a range of AOPs is available, photocatalysis and cavitation present promising options, particularly due to their ability to generate highly reactive oxidizing species without using additional chemicals such as peroxide or ozone. Hydrodynamic cavitation (HC), on the other hand, relies on the local change in pressure in a flowing fluid to generate and collapse vaporous microbubbles.134 HC could be an attractive choice for degrading microplastics by exploiting the shear intensity induced by bubble collapse. The sheer intensity of HC, in addition to its oxidizing radicals production, has been harnessed previously for biomass135 and solubilizing chemical oxygen demand (COD) even at typical industrial flow rates.136 It is hence possible to harness HC on its own or in synergy137–139 with other AOPs such as photocatalysis to eliminate the toxicity associated with microplastics in water, particularly in remote areas.Recommendations
There is an urgent need for a strategic combination of bioremediation coupled with a photocatalytic approach to eliminate MPs from the environment. Awareness needs to be spread among the public about the health hazards of plastic pollution, and stringent policies need to be amended to control plastic pollution in the environment.Author contributions
GPP and SP conceived the idea, acquired funding, and wrote, reviewed and edited the manuscript. KTU, VC, ND, AD, SN, AJE, MK drafted the manuscript. SN, AJE, and MK, helped with the discussion, and proofreading.Conflicts of interest
There are no conflicts to declare.Acknowledgements
KTU gratefully acknowledges SRM Institute of Science and Technology, Kattankulathur (Campus), Tamil Nadu, India for providing doctoral fellowship assistance (Ref: RA2223001011001). SP thanks Heriot-Watt University for start-up grant support.Notes and references
- J. N. Hitchcock and S. M. Mitrovic, Environ. Pollut., 2019, 247, 457–466 CrossRef CAS PubMed.
- M. Kedzierski, B. Lechat, O. Sire, G. Le Maguer, V. Le Tilly and S. Bruzaud, Food Packag. Shelf Life, 2020, 24, 100489 CrossRef.
- K. N. Palansooriya, M. K. Sang, A. D. Igalavithana, M. Zhang, D. Hou, P. Oleszczuk, J. Sung and Y. S. Ok, Environ. Res., 2022, 209, 112807 CrossRef CAS PubMed.
- Q. Yu, X. Hu, B. Yang, G. Zhang, J. Wang and W. Ling, Chemosphere, 2020, 249, 126059 CrossRef CAS PubMed.
- K. Karkanorachaki, P. Tsiota, G. Dasenakis, E. Syranidou and N. Kalogerakis, Microplastics, 2022, 1, 85–101 CrossRef.
- J. C. Prata, A. L. P. Silva, J. P. Da Costa, C. Mouneyrac, T. R. Walker, A. C. Duarte and T. Rocha-Santos, Int. J. Environ. Res. Public Health, 2019, 16, 2411 CrossRef PubMed.
- C. Sorasan, F. E. Ortega-Ojeda, A. Rodríguez and R. Rosal, Microplastics, 2022, 1, 198–210 CrossRef.
- N.-X. Geilfus, K. Munson, J. Sousa, Y. Germanov, S. Bhugaloo, D. Babb and F. Wang, Mar. Pollut. Bull., 2019, 145, 463–473 CrossRef CAS PubMed.
- S. Veerasingam, M. Ranjani, R. Venkatachalapathy, A. Bagaev, V. Mukhanov, D. Litvinyuk, L. Verzhevskaia, L. Guganathan and P. Vethamony, TrAC, Trends Anal. Chem., 2020, 133, 116071 CrossRef CAS.
- R. Karthik, R. Robin, R. Purvaja, D. Ganguly, I. Anandavelu, R. Raghuraman, G. Hariharan, A. Ramakrishna and R. Ramesh, Sci. Total Environ., 2018, 645, 1388–1399 CrossRef CAS PubMed.
- K. D. Cox, G. A. Covernton, H. L. Davies, J. F. Dower, F. Juanes and S. E. Dudas, Environ. Sci. Technol., 2019, 53, 7068–7074 CrossRef CAS PubMed.
- L. C. Jenner, J. M. Rotchell, R. T. Bennett, M. Cowen, V. Tentzeris and L. R. Sadofsky, Sci. Total Environ., 2022, 831, 154907 CrossRef CAS PubMed.
- R. Kumar, C. Manna, S. Padha, A. Verma, P. Sharma, A. Dhar, A. Ghosh and P. J. C. Bhattacharya, Chemosphere, 2022, 298, 134267 CrossRef CAS PubMed.
- J. Domenech, B. Annangi, R. Marcos, A. Hernández and J. Catalán, Mutat. Res., Rev. Mutat. Res., 2023, 108453 CrossRef CAS PubMed.
- P. Wu, S. Lin, G. Cao, J. Wu, H. Jin, C. Wang, M. H. Wong, Z. Yang and Z. Cai, J. Hazard. Mater., 2022, 129361 CrossRef CAS PubMed.
- Y. S. Ibrahim, S. Tuan Anuar, A. A. Azmi, W. M. A. Wan Mohd Khalik, S. Lehata, S. R. Hamzah, D. Ismail, Z. F. Ma, A. Dzulkarnaen and Z. Zakaria, JGH Open, 2021, 5, 116–121 CrossRef PubMed.
- W. Sun, S. Yan, Z. Meng, S. Tian, M. Jia, S. Huang, Y. Wang, Z. Zhou, J. Diao and W. Zhu, Environ. Int., 2022, 166, 107391 CrossRef CAS PubMed.
- R. Jiang, G. Lu, Z. Yan, J. Liu, D. Wu and Y. Wang, J. Hazard. Mater., 2021, 405, 124247 CrossRef CAS PubMed.
- S. P. Singh, S. M. Maliyekkal, T. Gupta and A. K. Agarwal, New Trends in Emerging Environmental Contaminants, 2022, pp. 3–8 Search PubMed.
- D. Chettri, T. Pati and A. K. Verma, Water Environ. J., 2023 DOI:10.1111/wej.12882.
- S. P. Singh, P. Sharma, A. Bano, A. K. Nadda and S. Varjani, Green Anal. Chem., 2022, 100030 CrossRef.
- A. L. Andrady, Mar. Pollut. Bull., 2017, 119, 12–22 CrossRef CAS PubMed.
- J. Li, H. Liu and J. P. Chen, Water Res., 2018, 137, 362–374 CrossRef CAS PubMed.
- S. Oberbeckmann and M. Labrenz, Annu. Rev. Mar. Sci., 2020, 12, 209–232 CrossRef PubMed.
- M. Shen, G. Zeng, Y. Zhang, X. Wen, B. Song and W. Tang, Sci. Total Environ., 2019, 697, 134200 CrossRef CAS PubMed.
- M. E. Seeley, B. Song, R. Passie and R. C. Hale, Nat. Commun., 2020, 11, 2372 CrossRef CAS PubMed.
- X. Feng, Q. Wang, Y. Sun, S. Zhang and F. Wang, J. Hazard. Mater., 2022, 424, 127364 CrossRef CAS PubMed.
- Z. Zhang, W. Peng, C. Duan, X. Zhu, H. Wu, X. Zhang and L. Fang, Gondwana Res., 2022, 108, 91–101 CrossRef CAS.
- M. C. Rillig, A. A. de Souza Machado, A. Lehmann and U. Klümper, Environ. Chem., 2018, 16, 3–7 CrossRef PubMed.
- H. Liu, L. Yue, Y. Zhao, J. Li, Y. Fu, H. Deng, D. Feng, Q. Li, H. Yu and Y. Zhang, Sci. Total Environ., 2022, 848, 157790 CrossRef CAS PubMed.
- W. Li, Z. Wang, W. Li and Z. Li, Chemosphere, 2022, 307, 135836 CrossRef CAS PubMed.
- L. Rong, L. Zhao, L. Zhao, Z. Cheng, Y. Yao, C. Yuan, L. Wang and H. Sun, Sci. Total Environ., 2021, 773, 145640 CrossRef CAS PubMed.
- H. Yu, L. Wang, Y. Lin, W. Liu, D. Tuyiringire, Y. Jiao, L. Zhang, Q. Meng and Y. Zhang, Ecotoxicol. Environ. Saf., 2020, 194, 110378 CrossRef CAS PubMed.
- D. Zhu, Q.-L. Chen, X.-L. An, X.-R. Yang, P. Christie, X. Ke, L.-H. Wu and Y.-G. Zhu, Soil Biol. Biochem., 2018, 116, 302–310 CrossRef CAS.
- P. Schwabl, S. Köppel, P. Königshofer, T. Bucsics, M. Trauner, T. Reiberger and B. Liebmann, Ann. Intern. Med., 2019, 171, 453–457 CrossRef PubMed.
- S. Xie, R. Zhang, Z. Li, C. Liu, Y. Chen and Q. Yu, Environ. Res., 2023, 217, 114861 CrossRef CAS PubMed.
- P. R. Gibson, Gut, 2004, 53, 1724–1725 CrossRef CAS PubMed.
- X. Zhang, H. Wang, S. Peng, J. Kang, Z. Xie, R. Tang, Y. Xing, Y. He, H. Yuan and C. Xie, Front. Public Health, 2022, 3963 Search PubMed.
- X. Zhu, L. Jiang, Y. Tu, Y. Tian, G. Xu, D. Wu, A. Li and X. Xie, Sci. Total Environ., 2022, 808, 151892 CrossRef CAS PubMed.
- H. Shen, Z. Zhao, Z. Zhao, Y. Chen and L. Zhang, Int. J. Mol. Sci., 2022, 23, 594 CrossRef CAS PubMed.
- A. Weber, A. Schwiebs, H. Solhaug, J. Stenvik, A. M. Nilsen, M. Wagner, B. Relja and H. H. Radeke, Environ. Int., 2022, 163, 107173 CrossRef CAS PubMed.
- W. Ren, C. Zhang, X. Wang and J. Wang, Environ. Sci. Pollut. Res., 2022, 29, 77625–77634 CrossRef CAS PubMed.
- Z. Yan, Y. Liu, T. Zhang, F. Zhang, H. Ren and Y. Zhang, Environ. Sci. Technol., 2021, 56, 414–421 CrossRef PubMed.
- R. Fujimoto, K. H. Harada and M. Minata, Environ. Sci. Technol., 2022, 56, 12778 CrossRef CAS PubMed.
- G. Kyrila, A. Katsoulas, V. Schoretsaniti, A. Rigopoulos, E. Rizou, S. Doulgeridou, V. Sarli, V. Samanidou and M. Touraki, J. Hazard. Mater., 2021, 413, 125363 CrossRef CAS PubMed.
- J. Meng, B. Xu, F. Liu, W. Li, N. Sy, X. Zhou and B. Yan, Chemosphere, 2021, 283, 131274 CrossRef CAS PubMed.
- S. Oberbeckmann, M. G. Loeder, G. Gerdts and A. M. Osborn, FEMS Microbiol. Ecol., 2014, 90, 478–492 CrossRef CAS PubMed.
- A. F. Ramsperger, A. C. Stellwag, A. Caspari, A. Fery, T. Lueders, H. Kress, M. G. Löder and C. Laforsch, Water, 2020, 12, 3216 CrossRef CAS.
- C. A. Medriano and S. Bae, Ecotoxicol. Environ. Saf., 2022, 245, 114125 CrossRef CAS PubMed.
- J.-N. Huang, B. Wen, J.-G. Zhu, Y.-S. Zhang, J.-Z. Gao and Z.-Z. Chen, Sci. Total Environ., 2020, 733, 138929 CrossRef CAS PubMed.
- X. Chen, J. Zhuang, Q. Chen, L. Xu, X. Yue and D. Qiao, Ecotoxicol. Environ. Saf., 2022, 241, 113809 CrossRef CAS PubMed.
- E. Fournier, J. Ratel, S. Denis, M. Leveque, P. Ruiz, C. Mazal, F. Amiard, M. Edely, V. Bezirard and E. Gaultier, J. Hazard. Mater., 2023, 443, 130383 CrossRef CAS PubMed.
- K. Kesy, M. Labrenz, B. S. Scales, B. Kreikemeyer and S. Oberbeckmann, Microorganisms, 2020, 9, 76 CrossRef PubMed.
- I. V. Kirstein, S. Kirmizi, A. Wichels, A. Garin-Fernandez, R. Erler, M. Löder and G. Gerdts, Mar. Environ. Res., 2016, 120, 1–8 CrossRef CAS PubMed.
- P. Jiang, S. Zhao, L. Zhu and D. Li, Sci. Total Environ., 2018, 624, 48–54 CrossRef CAS PubMed.
- L. Murphy, K. Germaine, D. N. Dowling, T. Kakouli-Duarte and J. Cleary, Proceedings of the 2nd International Conference on Microplastic Pollution in the Mediterranean Sea, Springer Cham, 2020, DOI:10.1007/978-3-030-45909-3.
- A. Tang, X. Bi, J. Du, L. Rao, V. Vasanthakumar, Y. B. Hu, M. L. Fu, W. Sun and B. Yuan, Sci. Total Environ., 2022, 834, 155322 CrossRef CAS PubMed.
- D. Hou, M. Hong, Y. Wang, P. Dong, H. Cheng, H. Yan, Z. Yao, D. Li, K. Wang and D. Zhang, Microorganisms, 2021, 9, 1909 CrossRef CAS PubMed.
- V. Moresco, A. Charatzidou, D. M. Oliver, M. Weidmann, S. Matallana-Surget and R. S. Quilliam, Environ. Pollut., 2022, 308, 119594 CrossRef CAS PubMed.
- M. Uribe, C. Hermosilla, A. Rodríguez-Durán, J. Vélez, S. López-Osorio, J. J. Chaparro-Gutiérrez and J. A. Cortés-Vecino, Pathogens, 2021, 10(9), 1152 CrossRef CAS PubMed.
- E. Zhang, M. Kim, L. Rueda, C. Rochman, E. VanWormer, J. Moore and K. Shapiro, Sci. Rep., 2022, 12, 6532 CrossRef CAS PubMed.
- L. Capurso, J. Clin. Gastroenterol., 2019, 53, S1–S41 CrossRef CAS PubMed.
- W. M. de Vos, H. Tilg, M. Van Hul and P. D. Cani, Gut, 2022, 71, 1020–1032 CrossRef CAS PubMed.
- P. Manasian, A.-S. Bustos, B. Pålsson, A. Håkansson, J. M. Peñarrieta, L. Nilsson and J. A. Linares-Pastén, Front. Microbiol., 2020, 11, 1534 CrossRef PubMed.
- H. Sharma and D. K. Neelam, J. Basic Microbiol., 2023, 63, 292–307 CrossRef CAS PubMed.
- A. Kumar, K. Dhar, S. S. Kanwar and P. K. Arora, Biol. Proced. Online, 2016, 18, 2 CrossRef PubMed.
- S. Chimalapati, M. de Souza Santos, A. E. Lafrance, A. Ray, W. R. Lee, G. Rivera-Cancel, G. Vale, K. Pawlowski, M. A. Mitsche, J. G. McDonald, J. Liou and K. Orth, Elife, 2020, 9 DOI:10.7554/eLife.58057.
- J. Jutkina, N. P. Marathe, C. F. Flach and D. G. J. Larsson, Sci. Total Environ., 2018, 616–617, 172–178 CrossRef CAS PubMed.
- Y. Sun, N. Cao, C. Duan, Q. Wang, C. Ding and J. Wang, J. Hazard. Mater., 2021, 409, 124979 CrossRef CAS PubMed.
- X. Yu, Z.-C. Zhou, X.-y. Shuai, Z.-j. Lin, Z. Liu, J.-y. Zhou, Y.-h. Lin, G.-s. Zeng, Z.-y. Ge and H. Chen, J. Hazard. Mater., 2023, 451, 131130 CrossRef CAS PubMed.
- K. Abe, N. Nomura and S. Suzuki, FEMS Microbiol. Ecol., 2020, 96(5) DOI:10.1093/femsec/fiaa031.
- X. Yan, X. Yang, Z. Tang, J. Fu, F. Chen, Y. Zhao, L. Ruan and Y. Yang, Environ. Pollut., 2020, 262, 114270 CrossRef CAS PubMed.
- R. Song, Y. Sun, X. Li, C. Ding, Y. Huang, X. Du and J. Wang, Sci. Total Environ., 2022, 828, 154596 CrossRef CAS PubMed.
- C. Xu, J. Lu, C. Shen, J. Wang and F. Li, Water Res., 2022, 225, 119192 CrossRef CAS PubMed.
- Q. Zeng, J. Xiang, C. Yang, J. Wu, Y. Li, Y. Sun, Q. Liu, S. Shi and Z. Gong, Chem. Eng. J., 2023, 454, 140193 CrossRef CAS.
- Y. Zhang, Y. A. Su, X. Qiu, Q. Mao, H. Liu, H. Liu, D. Wen and Z. Su, Bioresour. Technol., 2022, 364, 128005 CrossRef CAS PubMed.
- X. Tang, M. Zhou, G. Zeng and C. Fan, Chem. Eng. J., 2022, 435, 134734 CrossRef CAS.
- S. Wang, D. Zeng, B. Jin, Y. Su and Y. Zhang, Chem. Eng. J., 2023, 452, 139520 CrossRef CAS.
- Q. Yuan, R. Sun, P. Yu, Y. Cheng, W. Wu, J. Bao and P. J. J. Alvarez, J. Hazard. Mater., 2022, 427, 127895 CrossRef CAS PubMed.
- Z. Wang, J. Gao, H. Dai, Y. Zhao, D. Li, W. Duan and Y. Guo, J. Hazard. Mater., 2021, 409, 124981 CrossRef CAS PubMed.
- L. Xiaowei, F. Fang, L. Lanlan, D. Chengxun, H. Shuheng and Y. Zhimin, Asian J. Ecotoxicol., 2022, 200–210 Search PubMed.
- R. Kaur, A. Kumari, G. Sharma, D. Singh and R. Kaur, J. Appl. Microbiol., 2021, 131, 1274–1288 CrossRef CAS PubMed.
- Y. Yan, F. Zhu, C. Zhu, Z. Chen, S. Liu, C. Wang and C. Gu, Water Res., 2021, 204, 117597 CrossRef CAS PubMed.
- Z.-D. Wen, D.-W. Gao and W.-M. Wu, Appl. Microbiol. Biotechnol., 2014, 98, 4683–4690 CrossRef CAS PubMed.
- H.-M. Zhao, H. Du, J. Lin, X.-B. Chen, Y.-W. Li, H. Li, Q.-Y. Cai, C.-H. Mo, H.-M. Qin and M.-H. Wong, Sci. Total Environ., 2016, 562, 170–178 CrossRef CAS PubMed.
- J. B. Nshimiyimana, S. Khadka, P. Zou, S. Adhikari, R. Proshad, A. Thapa and L. Xiong, BMC Res. Notes, 2020, 13, 1–6 CrossRef PubMed.
- R. Nahurira, L. Ren, J. Song, Y. Jia, J. Wang, S. Fan, H. Wang and Y. Yan, Curr. Microbiol., 2017, 74, 309–319 CrossRef CAS PubMed.
- F. Li, Y. Liu, D. Wang, C. Zhang, Z. Yang, S. Lu and Y. Wang, PLoS One, 2018, 13, e0204324 CrossRef PubMed.
- N. Bai, S. Li, J. Zhang, H. Zhang, H. Zhang, X. Zheng and W. Lv, Environ. Pollut., 2020, 266, 115112 CrossRef CAS PubMed.
- P. Wang, J. Gao, Y. Zhao, M. Zhang and S. Zhou, Sci. Total Environ., 2021, 755, 142476 CrossRef CAS PubMed.
- J. Li, J. Zhang, M. P. Yadav and X. Li, Chemosphere, 2019, 225, 443–450 CrossRef CAS PubMed.
- A. Baker, B. Ahmad, K. M. Alarjani, N. saleem Aldosri and M. S. Khan, Chemosphere, 2021, 266, 128998 CrossRef CAS PubMed.
- M. Ahuactzin-Pérez, S. Tlecuitl-Beristain, J. García-Dávila, E. Santacruz-Juárez, M. González-Pérez, M. C. Gutiérrez-Ruíz and C. Sánchez, 3 Biotech, 2018, 8, 1–10 CrossRef PubMed.
- H. Yu, W. Qi, X. Cao, J. Hu, Y. Li, J. Peng, C. Hu and J. Qu, Environ. Int., 2021, 156, 106708 CrossRef CAS PubMed.
- C. Sun, G. Zhang, H. Zheng, N. Liu, M. Shi, X. Luo, L. Chen, F. Li and S. Hu, Aquat. Toxicol., 2019, 206, 81–90 CrossRef CAS PubMed.
- M. Ahuactzin-Pérez, S. Tlécuitl-Beristain, J. García-Dávila, E. Santacruz-Juárez, M. González-Pérez, M. C. Gutiérrez-Ruíz and C. Sánchez, Fungal Biol., 2018, 122, 991–997 CrossRef PubMed.
- D. Aksu, M. M. Diallo, U. Şahar, T. A. Uyaniker and G. Ozdemir, Arch. Microbiol., 2021, 203, 4101–4112 CrossRef CAS PubMed.
- P. Wang, Y. Zhang, J. Jin, T. Wang, J. Wang and B. Jiang, Sci. Total Environ., 2020, 746, 140455 CrossRef CAS PubMed.
- X. Huang, J. Shi, C. Cui, H. Yin, R. Zhang, X. Ma and X. Zhang, J. Appl. Microbiol., 2016, 121, 1616–1626 CrossRef CAS PubMed.
- A. Lara-Moreno, E. Morillo, F. Merchán and J. Villaverde, J. Environ. Manage., 2021, 289, 112512 CrossRef CAS PubMed.
- A. Kamyabi, H. Nouri and H. Moghimi, Ecotoxicol. Environ. Saf., 2018, 163, 471–477 CrossRef CAS PubMed.
- X. Peng, X. Qu, W. Luo and X. Jia, Bioresour. Technol., 2014, 169, 271–276 CrossRef CAS PubMed.
- X. Peng, Z. Zhang, W. Luo and X. Jia, Bioresour. Technol., 2013, 128, 173–179 CrossRef CAS PubMed.
- C. Hu, D. Huang, G. Zeng, M. Cheng, X. Gong, R. Wang, W. Xue, Z. Hu and Y. Liu, Chem. Eng. J., 2018, 338, 432–439 CrossRef CAS.
- C. Gu, J. Wang, Z. Zhao, Y. Han, M. Du, S. Zan and F. Wang, Environ. Sci. Pollut. Res., 2019, 26, 23832–23841 CrossRef CAS PubMed.
- Z. Asadgol, H. Forootanfar, S. Rezaei, A. H. Mahvi and M. A. Faramarzi, J. Environ. Health Sci. Eng., 2014, 12, 1–5 CrossRef PubMed.
- Y. Matsumura, A. Akahira-Moriya and M. Sasaki-Mori, Biocontrol Sci., 2015, 20, 35–42 CrossRef CAS PubMed.
- A. Fouda, A. Khalil, H. El-Sheikh, E. Abdel-Rhaman and A. Hashem, J. Adv. Biol. Biotechnol., 2015, 2, 123–132 CrossRef.
- A. Eltoukhy, Y. Jia, R. Nahurira, M. Abo-Kadoum, I. Khokhar, J. Wang and Y. Yan, BMC Microbiol., 2020, 20, 1–14 CrossRef PubMed.
- S. Miri, R. Saini, S. M. Davoodi, R. Pulicharla, S. K. Brar and S. Magdouli, Chemosphere, 2022, 286, 131670 CrossRef CAS PubMed.
- J. Ding, Y. Zhou, C. Wang, Z. Peng, Y. Mu, X. Tang and Z. Huang, Microb. Cell Fact., 2020, 19, 114 CrossRef CAS PubMed.
- K. H. D. Tang, S. S. M. Lock, P. S. Yap, K. W. Cheah, Y. H. Chan, C. L. Yiin, A. Z. E. Ku, A. C. M. Loy, B. L. F. Chin and Y. H. Chai, Sci. Total Environ., 2022, 832, 154868 CrossRef CAS PubMed.
- T. Yu, H. Ma, H. Zhang, M. Xiong, Y. Liu and F. Li, J. Environ. Manage., 2021, 288, 112450 CrossRef CAS PubMed.
- R. Ben Ali, S. Ben Ouada, C. Leboulanger, J. Ammar, S. Sayadi and H. Ben Ouada, Int. J. Phytorem., 2021, 23, 818–828 CrossRef CAS PubMed.
- Y. Jia, A. Eltoukhy, J. Wang, X. Li, T. S. Hlaing, M. M. Aung, M. T. Nwe, I. Lamraoui and Y. Yan, Int. J. Mol. Sci., 2020, 21, 3588–3593 CrossRef CAS PubMed.
- J. Sanchez-Aponte, I. Baldiris-Navarro, M. Torres-Virviescas and C. Bohorquez, Orient. J. Chem., 2019, 35, 1274 CAS.
- R. Mtibaà, D. R. Olicón-Hernández, C. Pozo, M. Nasri, T. Mechichi, J. González and E. Aranda, Ecotoxicol. Environ. Saf., 2018, 156, 87–96 CrossRef PubMed.
- F. S. de Santana, L. H. Gracioso, B. Karolski, M. dos Passos Galluzzi Baltazar, M. A. Mendes, C. A. O. do Nascimento and E. A. Perpetuo, Appl. Biochem. Biotechnol., 2019, 189, 103–115 CrossRef PubMed.
- L. Badiefar, B. Yakhchali, S. Rodriguez-Couto, A. Veloso, J. M. García-Arenzana, Y. Matsumura and M. Khodabandeh, RSC Adv., 2015, 5, 29563–29572 RSC.
- S. Zeng, J. Zhao and L. Xia, Bioprocess Biosyst. Eng., 2017, 40, 1237–1245 CrossRef CAS PubMed.
- Y. Wang, W. Zhang, Z. Zhang, W. Wang, S. Xu and X. He, J. Appl. Microbiol., 2021, 131, 208–220 CrossRef CAS PubMed.
- J. Yang, C. Guo, S. Liu, W. Liu, H. Wang, Z. Dang and G. Lu, Environ. Sci. Pollut. Res., 2018, 25, 17645–17653 CrossRef CAS PubMed.
- J. Li, F. Luo, D. Chu, H. Xuan and X. Dai, J. Basic Microbiol., 2017, 57, 941–949 CrossRef CAS PubMed.
- E. A. Perpetuo, E. C. N. da Silva, B. Karolski and C. A. O. do Nascimento, Biodegradation, 2020, 31, 331–340 CrossRef CAS PubMed.
- X. Zhang, L. Liu, S. Zhang, Y. Pan, J. Li, H. Pan, S. Xu and F. Luo, Biomed Res. Int., 2016, 2016, 5178697 Search PubMed.
- L. Feng, H. Liu, D. Cheng, X. Mao, Y. Wang, Z. Wu and Q. Wu, Biomed Res. Int., 2018, 2018, 3917054 Search PubMed.
- A. M. Nakamura, M. A. S. Kadowaki, A. Godoy, A. S. Nascimento and I. Polikarpov, Int. J. Biol. Macromol., 2018, 120, 1893–1905 CrossRef CAS PubMed.
- M. H. Fulekar, Bioresour. Bioprocess., 2017, 4, 28 CrossRef CAS PubMed.
- A. Expósito, D. Patterson, W. Mansor, J. Monteagudo, E. Emanuelsson, I. Sanmartín and A. Durán, J. Environ. Manage., 2017, 187, 504–512 CrossRef PubMed.
- A. J. Exposito, J. Monteagudo, A. Durán, I. San Martín and L. González, J. Hazard. Mater., 2018, 342, 597–605 CrossRef CAS PubMed.
- A. Durán, J. M. Monteagudo, D. Castillo and A. J. Expósito, J. Environ. Manage., 2022, 319, 115712 CrossRef PubMed.
- J. Monteagudo, A. Durán, R. González and A. J. Expósito, Appl. Catal. B, 2015, 176, 120–129 CrossRef.
- K. R. Davies, Y. Cherif, G. P. Pazhani, S. Anantharaj, H. Azzi, C. Terashima, A. Fujishima and S. Pitchaimuthu, J. Photochem. Photobiol., C, 2021, 48, 100437 CrossRef CAS.
- V. V. Ranade, V. M. Bhandari, S. Nagarajan, V. P. Sarvothaman and A. T. Simpson, Hydrodynamic Cavitation: Devices, Design and Applications, John Wiley & Sons, 2022 Search PubMed.
- S. Nagarajan and V. V. Ranade, Bioresour. Technol., 2022, 361, 127663 CrossRef CAS PubMed.
- S. Nagarajan and V. V. Ranade, Bioresour. Technol. Rep., 2020, 11, 100480 CrossRef.
- S. J. De-Nasri, V. P. Sarvothaman, S. Nagarajan, P. Manesiotis, P. K. Robertson and V. V. Ranade, Ultrason. Sonochem., 2022, 90, 106207 CrossRef CAS PubMed.
- S. J. De-Nasri, V. P. Sarvothaman, C. Kelly, S. Nagarajan, P. Manesiotis, P. K. Robertson and V. V. Ranade, Chem. Eng. J., 2023, 473, 145241 CrossRef CAS.
- V. P. Sarvothaman, V. K. Velisoju, J. Subburaj, M. S. Panithasan, S. R. Kulkarni, P. Castaño, J. Turner, P. Guida, W. L. Roberts and S. Nagarajan, Ultrason. Sonochem., 2023, 99, 106548 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |