Open Access Article
Open Access ArticleA copper(II) peptide helicate selectively cleaves DNA replication foci in mammalian cells†
Ana
Alcalde-Ordóñez‡
a,
Natalia
Barreiro-Piñeiro‡
b,
Bríonna
McGorman‡
c,
Jacobo
Gómez-González‡
a,
David
Bouzada
a,
Francisco
Rivadulla
 d,
M. Eugenio
Vázquez
d,
M. Eugenio
Vázquez
 a,
Andrew
Kellett
a,
Andrew
Kellett
 *c,
José
Martínez-Costas
*c,
José
Martínez-Costas
 *b and
Miguel Vázquez
López
*b and
Miguel Vázquez
López
 *e
*e
aCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Química Orgánica, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain
bCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Bioquímica e Bioloxía Molecular, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain
cSSPC, The SFI Research Centre for Pharmaceuticals, School of Chemical Sciences, Dublin City University, Glasnevin, Dublin 9, Ireland
dCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Química Física, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain
eCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Química Inorgánica, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain. E-mail: miguel.vazquez.lopez@usc.es
First published on 27th October 2023
Abstract
The use of copper-based artificial nucleases as potential anticancer agents has been hampered by their poor selectivity in the oxidative DNA cleavage process. An alternative strategy to solve this problem is to design systems capable of selectively damaging noncanonical DNA structures that play crucial roles in the cell cycle. We designed an oligocationic CuII peptide helicate that selectively binds and cleaves DNA three-way junctions (3WJs) and induces oxidative DNA damage via a ROS-mediated pathway both in vitro and in cellulo, specifically at DNA replication foci of the cell nucleus, where this DNA structure is transiently generated. To our knowledge, this is the first example of a targeted chemical nuclease that can discriminate with high selectivity 3WJs from other forms of DNA both in vitro and in mammalian cells. Since the DNA replication process is deregulated in cancer cells, this approach may pave the way for the development of a new class of anticancer agents based on copper-based artificial nucleases.
Introduction
The discovery of the cytotoxic properties of cisplatin by Barnett Rosenberg in 19651 gave rise to the new field of canonical dsDNA-targeted metallodrugs for cancer therapy.2 More recently, as our knowledge of the structural polymorphism and biology of DNA expanded, the bioinorganic field has shifted towards targeting noncanonical DNA structures, including single-stranded DNA, i-motifs, G-quadruplexes (G4s), DNA triplexes and DNA junctions, such as three-way (3WJs) and four-way (Holliday) junctions, among others.3–13 Here, we present a CuII peptide helicate that selective recognises 3WJs both in vitro and at DNA replication foci in mammalian cells. This is the first time such specificity has been achieved.3WJs are the simplest branched DNA structures,14 consisting of symmetrical assemblies formed by three convergent dsDNA units that meet at a central point called the branch point, which forms a hydrophobic cavity approximately 12 Å in diameter that is defined by the terminal base pairs of each of the three B-DNA arms.15 Metallosupramolecular helicates are the most promising agents for the selective recognition of these noncanonical DNA structures.6–12 One of the key factors for their selectivity is the high complementarity between their shape and the trigonal symmetry of the branch point of the 3WJs. Indeed, this is a key structural feature of other 3WJ binders, such as triptycene derivatives,16C3-symmetric cationic azacryptands,17 self-assembled supramolecular FeII tetrahedral metallocages18 and three-fold symmetric tripeptides.19
3WJs participate in a number of key biological processes,20–22 and notably they are transiently formed in the replication fork during DNA replication.23 Therefore, 3WJ-binding ligands are expected to stall the replication fork and elicit genotoxic replication stress in rapidly dividing cells, such as tumoral ones.24,25 For these reasons, 3WJs are currently considered a promising target for the development of new anticancer drugs with unique therapeutic properties.26–31
On the other hand, artificial CuII metallonucleases have been investigated as nucleic acid targeting drug molecules,32,33 and there are several examples in the literature of CuII complexes34–36 with remarkable anticancer activity combined with lower off-target toxicity than classical platinum drugs.37 Copper nucleases produce reactive oxygen species (ROS) that can cleave dsDNA through two alternative mechanisms: hydrolysis of the phosphodiester backbone, or oxidative H-atom abstraction from the deoxyribose ring.38 Copper-based nucleases themselves have little or no sequence selectivity,39 which limits their therapeutic potential. Consequently extensive research has been performed to generate hybrid chemical nucleases that can target specific nucleic acid structures and sequences. This has previously involved the attachment of well-studied chemical nucleases to nucleic acid recognition moieties such as dsDNA groove binders,40–43 non canonical DNA binders (specifically triplex DNA44 and G-quadruplexes45) or sequence specific dsDNA targeting agents.46–49 However, to date, no chemical nuclease, copper-derived or not, specifically directed against 3WJs has been described.
In 2021, we reported the first example of a chemical compound that selectively recognizes 3WJs in cells.50 The oligocationic ligand precursor of the metal complex that exhibited such activity was equipped with six artificial 2,2′-bipyridine-derived metal-binding residues that can fold predictably in the presence of FeII ions giving rise to a stable, water soluble, chiral dinuclear peptide helicate. That FeII peptide helicate, ΛΛ-FeII2LLD (Scheme 1), showed high affinity and selectivity towards 3WJs compared with canonical DNA in vitro and, importantly, selectively labelled DNA replication foci in the cell nuclei.50 Inspired by this precedent, herein we describe a redox active CuII peptide helicate that exhibit highly precise nuclease activity towards 3WJs in vitro, and can selectively cleaves DNA replication foci in mammalian cells. To our knowledge, the system reported herein is the first example of a chemical nuclease that exhibits high cleavage selectivity for 3WJs from other forms of DNA both in vitro and in mammalian cells. Since cancer cells have a deregulated DNA replication process, this approach may lead to the development of a new class of anticancer agents based on copper-based artificial nucleases.
Results and discussion
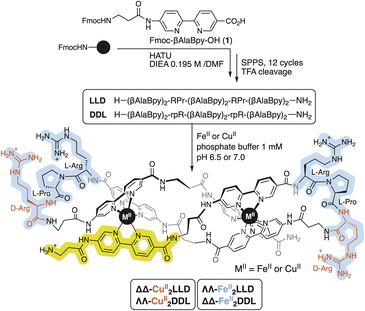
Synthesis and characterization of the CuII peptide helicate
The precursor peptide ligand LLD was assembled using standard Fmoc solid-phase peptide synthesis procedures.50 The Bpy ligand was introduced as an Fmoc-protected building block, Fmoc-βAlaBpy-OH (1; Scheme 1). The final peptide ligand was purified by HPLC, and its identity was confirmed by mass spectrometry (Fig. S1 and S5†).The formation in solution of the CuII2LLD peptide helicate (Scheme 1) was characterized by IR and UV spectroscopies (Fig. S3†) and by monitoring the quenching in the fluorescence emission band of the Bpy fluorophores present in LLD upon metal coordination (Fig. 1a and S4†). The resulting titration profile was fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (LLD
2 (LLD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CuII) model with a global dissociation constant of approximately 34.3 (9.4) nM. MALDI spectra of the solution at saturating concentrations of CuII ions showed a peak at m/z = 2555.97, consistent with the formation of the CuII2LLD peptide helicate (Fig. S4†).
CuII) model with a global dissociation constant of approximately 34.3 (9.4) nM. MALDI spectra of the solution at saturating concentrations of CuII ions showed a peak at m/z = 2555.97, consistent with the formation of the CuII2LLD peptide helicate (Fig. S4†).
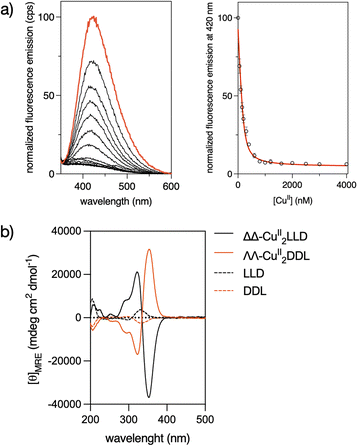
Circular Dichroism studies revealed a spectrum for the LLD peptide ligand that was dominated by a positive Cotton effect band at ca. 320 nm. This indicated an important preorganization of the peptide chain and the effective chiral induction of the heterochiral β-turn loops in combination with π-stacking interactions between the bipyridine units. Interestingly, the addition of CuII ions to a solution of LLD resulted in an inverted Cotton effect at this wavelength, accompanied with a large increase in its intensity. This is consistent with the formation of a ΔΔ- (or P) helicate (Fig. 1b). Surprisingly, this is the opposite chirality to that induced by FeII ions with the same LLD peptide ligand, which induce the formation of the diasteromeric (M) helicate.50 We also synthesised the isomeric CuII2DDL peptide helicate (Scheme 1), which, as expected, showed a mirror image CD spectrum to that of CuII2LLD, consistent with a ΛΛ configuration of the CuII metallopeptide (i.e., with the formation of a M helicate) (Fig. 1b).
It is known that the d9 configuration of CuII complexes results in pronounced Jahn–Teller distortion for six-coordinate systems and subsequent kinetic lability of axial ligands, even in chelated systems.51,52 It appears that the differences in the coordination behavior of FeII and CuII lead to different arrangements of the peptide ligand around the metal centres, leading to opposite helical chirality in the corresponding peptide helicates. To better define the coordination of CuII ions in the helicates, we performed EPR spectra at low (120 K) and room temperature (300 K) using a 200 μM solution of ΔΔ-CuII2LLD in phosphate buffer (Fig. S6†). At low temperature (i.e., frozen solution), the metallo-peptide spectra displayed a relatively symmetric signal, with an isotropic g = 2.197 that suggests regular coordination, indicative of octahedral or trigonal prismatic environments. In contrast, at room temperature, the narrowing of the lines suggests a possible rhombic distortion (Fig. S6†), as observed in other CuII trisbidentate octahedral complexes due to Jahn–Teller distortion.53
CuII peptide helicates recognize three-way DNA junctions in vitro
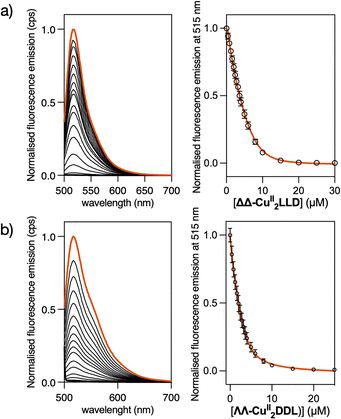
To study the DNA binding properties of the new CuII peptide helicates, we used a fluorescein-labelled 3WJ (Flu-3WJ; see ESI† Section 2.1 for DNA sequences) and measured its emission in the presence of increasing concentrations of the preformed ΔΔ-CuII2LLD or ΛΛ-CuII2DDL helicates. The progressive quenching of the fluorescein emission at 515 nm could be fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding mode (Flu-3WJ/helicate) plus nonspecific interactions, with an apparent dissociation constant KD = 260 nM for ΔΔ-CuII2LLD and KD = 654 nM for ΔΔ-CuII2DDL (Fig. 2). This indicates that the P helicate, ΔΔ-CuII2LLD, has higher affinity for the 3WJ. We also attempted to investigate the KD of the interaction of ΔΔ-CuII2LLD with fluorescein-labelled dsDNA (Flu-dsDNA), but the mixture precipitated during the titration, which suggests a nonspecific interaction between the peptide helicate and dsDNA.
1 binding mode (Flu-3WJ/helicate) plus nonspecific interactions, with an apparent dissociation constant KD = 260 nM for ΔΔ-CuII2LLD and KD = 654 nM for ΔΔ-CuII2DDL (Fig. 2). This indicates that the P helicate, ΔΔ-CuII2LLD, has higher affinity for the 3WJ. We also attempted to investigate the KD of the interaction of ΔΔ-CuII2LLD with fluorescein-labelled dsDNA (Flu-dsDNA), but the mixture precipitated during the titration, which suggests a nonspecific interaction between the peptide helicate and dsDNA.
To ensure that the ΔΔ-CuII2LLD could bind with 3WJs of different sequence compositions (i.e. different GC content) the fluorescein emission assay was repeated with a GC-rich 3WJ sequence (Flu-GC-3WJ, see ESI Section 2.1† for DNA sequences). A KD of 300 nM was obtained for the interaction between ΔΔ-CuII2LLD and Flu-GC-3WJ (Fig. S7†), which indicates that the affinity of this CuII peptide helicate for 3WJs is not limited to a single DNA sequence. The key role played by metal ions in the recognition of 3WJ by these family of peptide helicates was demonstrated in our previous work. There, we demonstrated by means of fluorescence anisotropy experiments the absence of interaction between the free peptide ligand LLD and 3WJ.50
Kinetic inertness of the ΔΔ-CuII peptide helicate
Next, we investigated the stability of CuII peptide helicate in solution, through a competitive binding assay with EDTA—an organic ligand that readily forms complexes with copper ions with extraordinary thermodynamic stability.54 Specifically, five equivalents of CuII ions were added to a 2 μM solution of LLD ligand, and fluorescence spectra were recorded before and after the addition of the metal ions, that is, before and after the assembly of the peptide helicate ΔΔ-CuII2LLD. Then, two equivalents of EDTA (with respect to the LLD peptide ligand) were added and the fluorescence spectra were recorded for 60 min. The experiment was repeated in the presence of 3WJ. It is necessary to clarify that the peptide ligand LLD is able to coordinate two CuII ions while the EDTA molecule is only able to coordinate a single metal ion, so to perform this classical competition study the stoichiometric ratio between the two ligands must be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (LLD
2 (LLD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDTA) against a fixed amount of CuII ions. The data obtained showed that the ligand substitution reaction takes ≥5 minutes to occur in the absence of 3WJ, but it slows to ≥20 minutes in the presence of 3WJ, which indicates that ΔΔ-CuII2LLD possesses, in addition to its high thermodynamic stability, a remarkable kinetic inertness in water media and probably also in cellulo (Fig. S8†).
EDTA) against a fixed amount of CuII ions. The data obtained showed that the ligand substitution reaction takes ≥5 minutes to occur in the absence of 3WJ, but it slows to ≥20 minutes in the presence of 3WJ, which indicates that ΔΔ-CuII2LLD possesses, in addition to its high thermodynamic stability, a remarkable kinetic inertness in water media and probably also in cellulo (Fig. S8†).
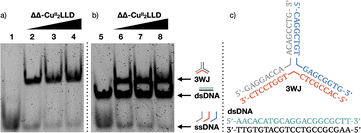
Analysis of three-way DNA junction recognition of the ΔΔ-CuII peptide helicate by PAGE
To further analyse the interaction of ΔΔ-CuII2LLD with 3WJ and dsDNA, we performed polyacrylamide gel electrophoresis (PAGE) experiments (Fig. 3a).55–57 In agreement with the fluorescence studies, we observed that incubation of 3WJ with increasing concentrations of preformed ΔΔ-CuII2LLD resulted in the appearance of a slower-migrating band consistent with the formation of the 3WJ/ΔΔ-CuII2LLD adduct (Fig. 3b). Here, almost all the 3WJ has been complexed with ΔΔ-CuII2LLD, even at the lowest concentration of helicate tested, which is expected for a high-affinity interaction. The intensity of this 3WJ band increases with the concentration of the CuII helicate (Fig. S9†), while the intensity of the single-stranded DNA band decreases. In contrast, no changes in the position or intensity of the dsDNA were observed upon incubation of the preformed ΔΔ-CuII2LLD helicate with dsDNA in the presence of 3WJ, confirming the low affinity this metallopeptide has for canonical dsDNA (Fig. 3b).The ΔΔ-CuII peptide helicate selectively cleaves three-way DNA junctions in vitro
To provide further clarity on 3WJ formation and enable ease of visualisation of target 3WJ and off-target dsDNA, PAGE experiments were also performed with fluorophore labelled sequences (Fig. 4a).58 3WJ strands were labelled at their 5′ end with FAM, ROX or Cy5, while the dsDNA strands were labelled with FAM and Cy5. Fluorophore 3WJ and dsDNA strands were then incubated with ΔΔ-CuII2LLD and visualised using a multiplex fluorescent assay (Fig. 4a). The blue (FAM), green (ROX) and red (Cy5) signal of the 3WJ sequences were combined to produce a yellow 3WJ band, while the blue (FAM) and red (Cy5) dsDNA strands appear pink. This multiplex binding assay was next applied for the analysis of the cleavage profile of ΔΔ-CuII2LLD.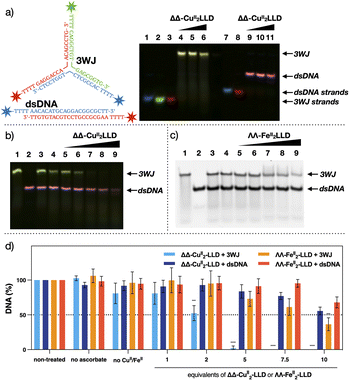 | ||
| Fig. 4 (a) Fluorescently labelled target 3WJ and off-target dsDNA. Blue = FAM, Green = ROX and Red = Cy5 (see ESI Section 2.3† for DNA sequences and experimental conditions). Multiplex image from 20% native analysis of the formation of 3WJ in the presence of increasing concentrations of ΔΔ-CuII2LLD. Lane 1: FAM-labelled single strand (ss) of 3WJ. Lane 2: ROX-labelled ss. Lane 3: Cy5-labelled ss of 3WJ. Lane 4–6: 3WJ strands incubated with 1, 2 or 3 eq. of ΔΔ-CuII2LLD, respectively. Lane 7: FAM-labelled ss of dsDNA. Lane 8: Cy5-labelled ss of dsDNA. Lanes 9–11: dsDNA incubated with 1, 2 or 3 eq. of ΔΔ-CuII2LLD, respectively. See Fig. S10† for individual images from multiplex assay. (b) PAGE analysis of the DNA damage inflicted by ΔΔ-CuII2LLD. Fluorophore labelled DNA was utilised to ensure clarity of target 3WJ DNA (yellow) and off-target dsDNA (pink). Lane 1: 3WJ. Lane 2; dsDNA. Lane 3: 3WJ and dsDNA incubated with ΔΔ-CuII2LLD. Lane 4: Target and off-target with LLD peptide ligand (no CuII present). Lanes 5–9: 3WJ and dsDNA incubated with 1, 2, 5, 7.5 or 10 eq. of ΔΔ-CuII2LLD and Na-L-ascorbate (10 eq. to ΔΔ-CuII2LLD). Individual images available in Fig. S11.† (c) Control cleavage experiment performed with ΛΛ-FeII2LLD. Lane 1: 3WJ DNA. Lane 2: dsDNA off-target. Lane 3; control of 3WJ and dsDNA incubated with ΛΛ-FeII2LLD. Lane 4: 3WJ and dsDNA with non-metalated LLD peptide ligand. Lanes 5–9: 3WJ and dsDNA incubated with 1, 2, 5, 7.5 or 10 eq. of ΛΛ-FeII2LLD and Na-L-ascorbate (10 eq. to ΛΛ-FeII2LLD). (d) Band densitometry analysis of cleavage induced upon 3WJ and dsDNA by ΔΔ-CuII2LLD (Fig. S12†) and ΛΛ-FeII2LLD (Fig. 4c and S13†). * = P ≤ 0.05 ** = P ≤ 0.01 *** = P ≤ 0.001 and **** = P ≤ 0.0001. Na-L-ascorbate concentration is 10 eq. to ΔΔ-CuII2LLD or ΛΛ-FeII2LLD. | ||
The ability of ΔΔ-CuII2LLD to selectively recognise and cleave 3WJs in vitro was analysed by PAGE (Fig. 4b). The fluorescently labelled target 3WJ and off-target dsDNA oligos (see ESI 2.3† for DNA sequences) were incubated with increasing equivalents ΔΔ-CuII2LLD and Na-L-ascorbate at 37 °C for 24 hours. PAGE analysis revealed that the CuII peptide helicate selectively cleaves 3WJ, which fully disappears when exposed to 5 eq. of ΔΔ-CuII2LLD, while the dsDNA present in the sample remains intact when exposed up to 7.5 equivalents of the CuII peptide helicate. A control cleavage experiment was also performed with ΛΛ-FeII2LLD. Here, 3WJ and dsDNA were incubated with the FeII helicate and Na-L-ascorbate under identical conditions to that of the ΔΔ-CuII2LLD assay, and the cleavage profile was visualised by PAGE (Fig. 4c). Band densitometry revealed a significant difference between the cleavage ΔΔ-CuII2LLD induced towards 3WJ compared to the off-target dsDNA; for example, just 2% of the 3WJ remained versus 80% of off-target dsDNA following exposure to 5 eq. of ΔΔ-CuII2LLD (Fig. 4d and S14†). By contrast, ΛΛ-FeII2LLD did not show selectivity in the cleavage of 3WJ versus dsDNA across all tested concentrations (Fig. 4d and S14†). Moreover, the FeII peptide helicate is only able to cleave 70% of 3WJ at the highest concentration of metallopeptide tested (10 eq.), which highlights its lack of efficiency.
A time point study was performed to determine the incubation period required to achieve 3WJ selective damage (Fig. S14†). This revealed that cleavage initiates within 1 hour and increases up to 12 hours; from 12 to 24 hours no significant increase in cleavage was observed. This indicates that ΔΔ-CuII2LLD can rapidly and selectively damage DNA 3WJs, and that off-target dsDNA damage would not be expected upon longer incubation. Combining the results of Fig. 3, 4 and S10–S15,† it can be concluded that ΔΔ-CuII2LLD can discriminate with great efficiency between 3WJ and dsDNA, and selectively cleave 3WJ in the presence of canonical DNA. To our knowledge, this is the first example of such activity for a chemical nuclease in vitro.
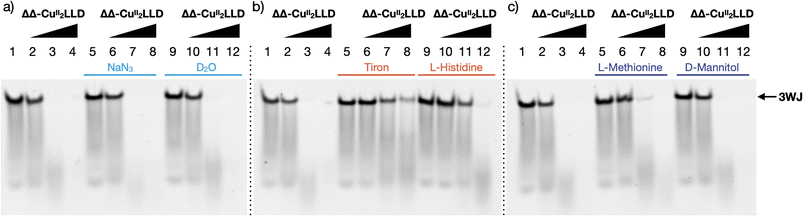
Next, the cleavage mechanism for the ΔΔ-CuII2LLD helicate was investigated using reactive oxygen species (ROS) scavengers.59,60 Here, 3WJ was incubated with ΔΔ-CuII2LLD, Na-L-ascorbate, and a series of selective ROS scavengers (L-histidine, D-mannitol, L-methionine, and tiron (sodium catechol sulfate)), and the resulting cleavage profiles were visualized by PAGE (Fig. 5). The presence of tiron significantly impeded the activity of the peptide helicate, indicating that superoxide radicals (O2˙−)61 play a dominant role in the 3WJ cleavage inflicted by ΔΔ-CuII2LLD. To probe this finding further, we determined if diffusible superoxide radicals were directly contributing to DNA cleavage by examining the activity in the presence of 4-hydroxy-TEMPO, a known scavenger of difussible O2˙− (Fig. S16†).62 TEMPO resulted in negligible cleavage inhibition indicating that difussible O2˙− plays a minimal role in ΔΔ-CuII2LLD cleavage and that, instead, oxidative damage is likely mediated by a copper bound superoxo radical species (Cu–O2˙−) proximate to the 3WJ. The scavenger L-histidine also impeded 3WJ damage, which suggested that singlet oxygen (1O2) is contributing to the cleavage mechanism. However, since 1O2 is usually generated under photoactivated conditions we further investigated the role of this ROS in ΔΔ-CuII2LLD activity. The scavenger assay was therefore expanded to include deuterium oxide (D2O)—a known 1O2 stabiliser—and sodium azide (NaN3)—a known scavenger of 1O2 (Fig. 5a). The presence of D2O and NaN3 did not perturb 3WJ cleavage and consequently it is not clear at this point how 1O2 mediates ΔΔ-CuII2LLD activity. It should be noted that several recent studies reporting the dsDNA damaging effects of polynuclear CuII metallonucleases have predicted 1O2-mediated DNA damage and identified L-histidine as viable scavenger in vitro and in cellulo.63,64
A rhodamine-labelled ΔΔ-CuII peptide helicate selective labels DNA-replication sites in mammalian cells
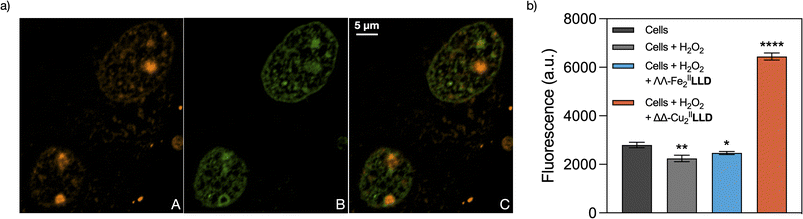
To this point we have demonstrated that the ΔΔ-CuII2LLD readily forms in solution, its stability in aqueous environments, and that ΔΔ-CuII2LLD can selectively bind and damage 3WJ in vitro. Next, in cellulo studies were performed to investigate the ability of the CuII helicate to selectively target DNA replication centres (i.e., 3WJs in the cell nuclei). Preliminary experiments indicated, as was the case with FeII analogues,50 that a TAMRA-labelled CuII peptide helicate, ΔΔ-CuII2TAMRA-LLD (Fig. S2†), did not internalise in Vero cells. However, it readily translocated in digitonin-treated cells where it showed a diffused distribution in the cytosol, and a strong punctuated distribution in the nucleus, as well as an evident concentration in the nucleolus (Fig. 6a(A)—red and Fig. S18†). Most of the nuclear spots co-localized with GFP-tagged Proliferating Nuclear Antigen (PCNA), which specifically labels DNA replication foci in actively dividing cells (Fig. 6a(B)—green).65 While its cytosolic distribution and association with the nucleoli might reflect some off-target RNA-binding of the helicate, replisomes labelled by PCNA are enriched in 3WJs due to the presence of replication forks. Thus, our results strongly suggest the direct association of the CuII peptide helicate with 3WJs structures in functional cell nuclei. Moreover, control experiments show that the free LLD peptide ligand does not localise in the DNA replication centres (Fig. S17†). As already suggested by the results obtained in the competition studies with EDTA described above, this control experiment further demonstrates that the ΔΔ-CuII2LLD helicate is stable in cell medium and that it only binds DNA replication centres in the nucleus as a discrete metallopeptide.The ΔΔ-CuII peptide helicate selective cleaves DNA-replication foci in mammalian cells
Having demonstrated that ΔΔ-CuII2LLD is stable in cellulo and associates with 3WJ-enriched foci in cellular DNA, we next investigated if it could exert its nuclease activity upon chromatin-associated 3WJs in cells. To maximize the number of cells undergoing DNA replication, and thus increase the chances of detecting free DNA ends obtained by nuclease cleavage of 3WJs, we used Vero cells that were previously synchronised by 42 h of serum deprivation.66 16 h after re-supplementation of the cultures with 10% serum—when most of the cells were in S phase—the cells were pre-treated with digitonin and then incubated with H2O2 in the presence or absence of ΔΔ-CuII2LLD. Cells were then incubated for 2 h at 37 °C and subjected to a Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labelling (TUNEL) assay. The TUNEL assay uses TdT polymerase to label the free ends of DNA with a fluorescent dUTP, and thereby enables detection of cleavage sites. Here, the free DNA ends in the cell nuclei DNA (i.e., DNA sites cleaved by ΔΔ-CuII2LLD) were detected through Alexa Fluor 488™ fluorescence.67 Very importantly, the results show that incubation of ΔΔ-CuII2LLD in the presence of H2O2 produced a significant increase for the TUNEL fluorescent signal (Fig. 6b), which indicates the presence of free DNA ends and thus, nuclease activity in cellulo. A control experiment was also performed with ΛΛ-FeII2LLD in the presence of H2O2. Significantly, the increase in fluorescent signal—indicative of DNA cleavage—was not observed for the FeII helicate, which agrees with the above-described in vitro studies, which suggested a lack of nuclease activity. To our knowledge, this is the first example of such activity for a chemical nuclease in cellulo.Conclusion
We reported herein the synthesis of two new chiral dinuclear oligocationic CuII peptide helicates derived from two enantiomeric peptide ligands equipped with six chelating 2,2′-bipyridine residues, named LLD and DDL, following a SPPS methodology.50 Spectrofluorimetric and PAGE DNA binding studies indicate that both CuII peptide helicates can recognize 3WJs in vitro with high affinity and selectivity, even in the presence of canonical DNA, showing chiral discrimination between the two diastereoisomers, being ΔΔ-CuII2LLD the metallopeptide with higher 3WJ affinity of the two (KD = 260 nm vs. 654 nm). Moreover, the kinetic inertness of ΔΔ-CuII2LLD in water was investigated through a competition study with EDTA in presence and absence of 3WJ, suggesting that ΔΔ-CuII2LLD is very stable in vitro and in cellulo.The studies carried out clearly show that ΔΔ-CuII2LLD is an efficient and selective in vitro agent for the cleavage of 3WJs, even in the presence of canonical DNA. The mechanism through which this 3WJ cleavage by ΔΔ-CuII2LLD occurred was also investigated. Studies using ROS scavengers showed that metal-bound superoxide radicals (Cu–O2˙−) play the key role in the cleavage process and suggest that singlet oxygen (1O2) is also involved in some way.
The ability ΔΔ-CuII2LLD to internalise in cells was studied using of CuII helicate labelled with TAMRA (ΔΔ-CuII2TAMRA-LLD). This helicate was effectively internalised by digitonin-treated mammalian cells and was localised into the nucleus. Its nuclear distribution matched the localization of DNA replication sites, demonstrating that ΔΔ-CuII2LLD binds with high affinity and selectivity to 3WJ in cellulo. Finally, the selective cleavage of DNA replication foci in cells was evaluated by TUNEL assay. The studies performed indicate that ΔΔ-CuII2LLD produced a significant increase of the TUNEL fluorescent signal in the presence of H2O2, which indicates the presence of free DNA ends and thus, nuclease activity in cellulo.
To our knowledge, ΔΔ-CuII2LLD is the first example of a chemical nuclease that selectively cleaves 3WJ both in vitro and in cellulo. As DNA replication is deregulated in cancer cells, we strongly believe that this work will form the basis of a new class of anticancer metallodrug that target and damage with high efficiency and selectivity this noncanonical DNA structure that is transiently generated in the DNA replication foci of cell nuclei.
Data availability
Raw data for Fig. 1–6 are published in the figshare repository https://doi.org/10.6084/m9.figshare.22754600.Author contributions
M. V. L., J. M.-C. and A. K. conceived and designed the investigations. M. E. V. deeply assisted in the design of the investigations, interpretation of experimental data and proofreading of the manuscript. A. A.-O., N. B.-P., B. McG. and J. G.-G. performed the experiments. D. B. assisted in conducting DNA-binding studies. F. R. conducted the EPR studies. M. V. L, J. M.-C. and A. K. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
M. V. L., M. E. V., and J. M.-C. thank grants RTI2018-099877-B-I00, PID2021-127857NB-I00, PID2019-105308RB-I00 and PID2021-127702NB-I00 by MCIN/AEI/10.13039/501100011033 and by ERDF A way of making Europe. M. V. L. thanks Fundación Científica de la Asociación Española Contra el Cáncer (Ideas Semilla 2021 – IDEAS211154VÁZQ). M. V. L, J. M.-C, M. E. V and F. R. thank Xunta de Galicia (grants ED431C 2021/29 and ED431B 2021/13). J. G.-G. thanks the USC and NextGeneration EU program for his Margarita Salas postdoc fellowship. A. A.-O thanks the Spanish Ministry of Science and Innovation/Spanish Research Agency for her FPI fellowship. A. K. and B. McG. acknowledge funding from the Irish Research Council (EPSPD/2022/164 and IRCLA/2022/3815), Science Foundation Ireland funded research centers SSPC and CÚRAM (12/RC/2275 P2), and the European Union's Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement No. 861381.Notes and references
- R. A. Alderden, M. D. Hall and T. W. J. Hambley, The discovery and development of cisplatin, J. Chem. Educ., 2006, 83, 728–734 CrossRef CAS.
- E. J. Anthony, E. M. Bolinho, H. E. Bridgewater, O. W. L. Carter, J. M. Donnely, C. Imberti, E. C. Lant, F. Lermyte, R. J. Needham, M. Palau, P. J. Sadler, H. Shi, F.-X. Wang, W.-Y. Zhang and Z. Zhang, Metallodrugs are unique: opportunities and challenges of discovery and development, Chem. Sci., 2020, 11, 12888–12917 RSC.
- A. Shivaligam, M. A. Izquierdo, A. L. Marois, A. Vysniauskas, K. Suhling, M. K. Kuimova and R. Vilar, The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy, Nat. Commun., 2015, 6, 8178 CrossRef PubMed.
- R. Vilar, Interaction of metal complexes with G-quadruplex DNA, Adv. Inorg. Chem., 2020, 75, 425–445 CrossRef CAS.
- D. Bouzada, I. Salvadó, G. Barka, G. Rama, J. Martínez-Costas, R. Lorca, A. Somoza, M. Melle-Franco, M. E. Vázquez and M. Vázquez López, Selective G-quadruplex binding by oligoarginine-Ru(dppz) metallopeptides, Chem. Commun., 2018, 54, 658–661 RSC.
- A. Oleksi, A. G. Blanco, R. Boer, I. Usón, J. Aymamí, A. Rodger, M. J. Hannon and M. Coll, Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate, Angew. Chem., Int. Ed., 2006, 45, 1227–1231 CrossRef CAS PubMed.
- C. A. J. Hooper, L. Cardo, J. S. Craig, L. Melidis, A. Garai, R. T. Egan, V. Sadovnikova, F. Burkert, L. Male, N. J. Hodges, D. D. F. Browning, R. Rosas, F. Liu, F. V. Rocha, M. A. Lima, S. Liu, D. Bardelang and M. J. Hannon, Rotaxanating metallo-supramolecular nano-cylinder helicates to switch DNA junction binding, J. Am. Chem. Soc., 2020, 142, 20651–20660 CrossRef CAS PubMed.
- A. D. Faulkner, R. A. Kaner, Q. M. A. Abdallah, G. Clarkson, D. J. Fox, P. Gurnani, S. E. Howson, R. M. Phillips, D. I. Roper, D. H. Simpson and P. Scott, Asymmetric triplex metallohelices with high and selective activity against cancer cells, Nat. Chem., 2014, 6, 797–803 CrossRef CAS PubMed.
- H. Song, S. A. Allison, V. Brabec, H. E. Bridgewater, J. Kasparkova, H. Jostrhunova, V. Novohradsky, R. M. Phillips, J. Pracharova, N. J. Rogers, S. L. Shepherd and P. Scott, Glycoconjugated metallohelices have improved nuclear delivery and suppress tumour growth in vivo, Angew. Chem., Int. Ed., 2020, 59, 14677–14685 CrossRef CAS PubMed.
- J. Gómez-González, D. Bouzada, L. A. Pérez-Márquez, G. Sciortino, J.-D. Maréchal, M. Vázquez López and M. E. Vázquez, Stereoselective self-assembly of DNA binding helicates directed by the viral β-annulus trimeric peptide motif, Bioconjugate Chem., 2021, 32, 1564–1569 CrossRef PubMed.
- J. Gómez-González, D. G. Peña, G. Barka, G. Sciortino, J.-D. Maréchal, M. Vázquez López and M. E. Vázquez, Directed self-assembly of trimeric DNA-binding chiral miniprotein helicates, Front. Chem., 2018, 6, 520 CrossRef PubMed.
- I. Gamba, G. Rama, E. Ortega-Carrasco, J.-D. Maréchal, J. Martínez-Costas, M. E. Vázquez and M. Vázquez López, Programmed stereoselective assembly of DNA-binding helical metallopeptides, Chem. Commun., 2014, 50, 11097–11100 RSC.
- J. S. Craig, L. Melidis, H. D. Williams, S. J. Dettmer, A. A. Heidecker, P. J. Altmann, S. Guan, C. Campbell, D. F. Browning, R. K. O. Sigel, S. Johannsen, R. T. Egan, B. Aikman, A. Casini, A. Pöthig and M. J. Hannon, Organometallic pillarplexes that bind DNA 4-way Holliday junctions and forks, J. Am. Chem. Soc., 2023, 145, 13570–13580 CrossRef CAS PubMed.
- K. J. Neelsen and M. Lopes, Replication fork reversal in eukaryotes: from dead end to dynamic response, Nat. Rev. Mol. Cell Biol., 2015, 16, 207–220 CrossRef CAS PubMed.
- K. C. Woods, S. S. Martin, V. C. Chu and E. P. Baldwin, Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction, J. Mol. Biol., 2001, 313, 49–69 CrossRef CAS PubMed.
- S. A. Barros and D. M. Chenoweth, Recognition of nucleic acid junctions using triptycene-based molecules, Angew Chem. Int. Ed. Engl., 2014, 53, 13746–13750 CrossRef CAS PubMed.
- J. Novotna, A. Laguerre, A. Granzhan, M. Pirrotta, M.-P. Teulade-Fichou and D. Monchaud, Cationic azacryptands as selective three-way DNA junctions binding agents, Org. Biomol. Chem., 2015, 13, 215–222 RSC.
- J. Zhu, C. J. E. Haynes, M. Kieffer, J. L. Greenfield, R. D. Greenhalgh, J. R. Nitschke and U. F. Keyser, FeII4L4 tetrahedron binds to nonpaired DNA bases, J. Am. Chem. Soc., 2019, 141, 11358–11362 CrossRef CAS PubMed.
- J. Gómez-González, L. Martínez-Castro, J. Tolosa-Barrilero, A. Alcalde, S. Learte-Aymami, J. L. Mascareñas, J. C. García-Martínez, J. Martínez-Costas, J. D. Maréchal, M. Vázquez López and M. E. Vázquez, Selective recognition of A/T-rich DNA 3-way junctions with a three-fold symmetric tripeptide, Chem. Commun., 2022, 58, 7769–7772 RSC.
- J. Atkinson and P. McGlynn, Replication fork reversal and the maintenance of genome stability, Nucleic Acid Res., 2009, 37, 3475–3492 CrossRef CAS PubMed.
- L. S. Shlyakhtenko, V. N. Potaman, R. R. Sinden, A. A. Gall and Y. L. Lyubchenko, Structure and dynamics of three-way DNA junctions: atomic force microscopy studies, Nucleic Acid Res., 2000, 28, 3472–3477 CrossRef CAS PubMed.
- R. R. Sinden, M. J. Pytlos-Sinden and V. N. Potaman, Slipped strand DNA structures, Front. Biosci., 2007, 12, 4788–4799 CrossRef CAS PubMed.
- C.-H. Hu, F.-Y. Xu, K. Wang, A. N. Pearson and G. D. Pearson, Symmetrical adenovirus minichromosomes have hairpin replication intermediates, Gene, 1992, 110, 145–150 CrossRef CAS PubMed.
- H. Liao, F. Ji, T. Helleday and S. Ying, Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments, EMBO Rep., 2008, 19, e46263, DOI:10.15252/embr.201846263.
- K. Duskova, P. Lejault, É. Penchimol, R. Guillot, S. Britton, A. Granzhan and D. Monchaud, DNA junction ligands trigger DNA damage and are synthetic lethal with DNA repair inhibitors in cancer cells, J. Am. Chem. Soc., 2020, 142, 424–435 CrossRef CAS PubMed.
- A. Bacolla and R. D. Wells, Non-B DNA conformations as determinants of mutagenesis and human disease, Mol. Carcinog., 2009, 48, 273–285 CrossRef CAS PubMed.
- R. D. Wells, Non-B DNA conformations, mutagenesis and disease, Trends Biochem. Sci., 2007, 32, 271–278 CrossRef CAS PubMed.
- H. Tateishi-Karimata and N. Sugimoto, Roles of non-canonical structures of nucleic acids in cancer and neurodegenerative diseases, Nucleic Acid Res., 2021, 49, 7839–7855 CrossRef CAS PubMed.
- J. Zell, F. R. Sperti, S. Britton and D. Monchaud, DNA folds threaten genetic stability and can be leveraged for chemotherapy, RSC Chem. Biol., 2021, 2, 47–76 RSC.
- H. Tateishi-Karimata and N. Sugimoto, Chemical biology of non-canonical structures of nucleic acids for therapeutic applications, Chem. Commun., 2020, 56, 2379–2390 RSC.
- L. A. Howell and M. Searcey, Targeting higher-order DNA: beyond the G-quadruplex, ChemBioChem, 2009, 10, 2139–2143 CrossRef CAS PubMed.
- F. Mancin, P. Scrimin, P. Tecilla and U. Tonellato, Artificial metallonucleases, Chem. Commun., 2005, 2540–2548 RSC.
- L. J. Childs, J. Malina, B. E. Rolfsnes, M. Pascu, M. J. Prieto, M. J. Broome, P. M. Rodger, E. Sletten, V. Moreno, A. Rodger and M. J. Hannon, A DNA-binding copper(I) metallosupramolecular cylinder that acts as an artificial nuclease, Chem.–Eur. J., 2006, 12, 4919–4927 CrossRef CAS PubMed.
- Z.-R. Li, J. Li, W. Cai, J. Y. H. Lai, S. M. K. McKinnie, W.-P. Zhang, B. S. Moore, W. Zhang and P.-Y. Qian, Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage, Nat. Chem., 2019, 11, 880–889 CrossRef CAS PubMed.
- C. Wen-de, C. Lüdtke and M. Kulak, Copper complexes of N-donor ligands as artificial nucleases, Eur. J. Inorg. Chem., 2014, 2597–2612 CrossRef CAS.
- N. Z. Fantoni, Z. Molphy, S. O'Carroll, G. Menounou, G. Mitrikas, M. G. Krokidis, C. Chatgiliaglu, J. Colleran, A. Banasiak, M. Clynes, S. Roche, S. Kelly, V. McKee and A. Kellet, Polypyridyl-based copper phenanthrene complexes: combining stability with enhanced DNA recognition, Chem.–Eur. J., 2020, 27, 971–983 CrossRef PubMed.
- T. J. P. McGivern, S. Afsharpour and C. J. Marmion, Copper complexes as artificial DNA metallonucleases: From Sigman's reagent to next generation anti-cancer agent?, Inorg. Chim. Acta, 2018, 472, 12–39 CrossRef CAS.
- C. Liu, M. Wang, T. Zhang and H. Sun, DNA hydrolysis promoted by di- and multi-nuclear metal complexes, Coord. Chem. Rev., 2004, 248, 147–168 CrossRef CAS.
- K. J. Humphreys, K. D. Karlin and S. E. Kokita, Recognition and strand scission at junctions between single- and double-stranded DNA by a trinuclear copper complex, J. Am. Chem. Soc., 2001, 123, 5588–5589 CrossRef CAS PubMed.
- P. G. Shultz, J. S. Taylor and P. B. Dervan, Design synthesis of a sequence-specific DNA cleaving molecule. (Distamycin-EDTA)iron(II), J. Am. Chem. Soc., 1982, 104, 6861–6863 CrossRef CAS.
- M. Pitié, C. J. Burrows and B. Meunier, Mechanisms of DNA cleavage by copper complexes of 3-Clip-Phen and of its conjugate with a distamycin analogue, Nucleic Acid Res., 2000, 28, 4856–4864 CrossRef PubMed.
- C. H. B. Chen, A. Mazumber, J. F. Constant and D. S. Sigman, New conjugates with low molecular weight targeting ligands, Bioconjugate Chem., 1993, 4, 69–77 CrossRef CAS PubMed.
- M. Pitié, A. Croisy, D. Carrez, C. Blodron and B. Meunier, Cytostatic activity of 1,10-phenantroline derivatives generated by the clip-phen strategy, ChemBioChem, 2005, 6, 686–691 CrossRef PubMed.
- A. Zaid, J.-S. Sun, C.-H. Nguyen, E. Bisagni, T. Garestier, D. S. Grierson and R. Zain, Triple-helix directed cleavage of double-stranded DNA by benzoquinoquinozaline-1,10-phenantroline conjugates, ChemBioChem, 2004, 5, 1550–1557 CrossRef CAS PubMed.
- Z. Yu, M. Han and J. Cowan, Toward the design of a catalytic metallodrug: selective cleavage of G-quadruplex telomeric DNA by an anticancer copper-acridine-ATCUN complex, Angew. Chem., Int. Ed., 2015, 54, 1901–1905 CrossRef CAS PubMed.
- O. Luige, P. P. Bose, R. Stultz, P. Steunenberg, O. Brun, S. Andersson, M. Murtola and R. Strömberg, Zn2+-dependent peptide nucleic acid-based artificial ribonucleases with unprecedent efficiency and specificity, Chem. Commun., 2021, 57, 10911–10914 RSC.
- B. McGorman, N. Z. Fantoni, S. O'Carroll, A. Ziemele, A. H. El-Sagheer, T. Brown and A. Kellett, Enzymatic synthesis of chemical nuclease triplex-forming oligonucleotides with gene-silencing applications, Nucleic Acid Res., 2020, 50, 5467–5481 CrossRef PubMed.
- N. Z. Fantoni, B. McGorman, Z. Molphy, D. Singleton, S. Walsh, A. H. El-Sagheer, V. McKee, T. Brown and A. Kellett, Development of gene-targeted polypyridyl triplex-forming oligonucleotide hybrids, ChemBioChem, 2020, 21, 3563–3574 CrossRef PubMed.
- T. Lauria, C. Slator, V. McKee, M. Müller, S. Stazzoni, A. L. Crisp, T. Carell and A. Kellett, A click chemistry approach to developing molecularly targeted DNA scissors, Chem.–Eur. J., 2020, 26, 16782–16792 CrossRef CAS PubMed.
- J. Gómez-González, Y. Pérez, G. Sciortino, L. Roldan-Martín, J. Martínez-Costas, J.-D. Maréchal, I. Alfonso, M. Vázquez López and M. E. Vázquez, Dynamic stereoselection of peptide helicates and their selective labeling of DNA replication foci in cells, Angew. Chem., Int. Ed., 2021, 60, 8859–8866 CrossRef PubMed.
- D. H. Powell, L. Helm and A. E. Merbach, 17O nuclear magnetic resonance in aqueous solutions of Cu2+: the combined effect of Jahn–Teller inversion and solvent exchange on relaxation rates, J. Chem. Phys., 1991, 95, 9258–9265 CrossRef CAS.
- M. A. Halcrow, Jahn–Teller distortions in transition metal compounds, and their importance in functional molecular and inorganic materials, Chem. Soc. Rev., 2013, 42, 1784–1795 RSC.
- M. V. Veidis, G. H. Schreiber, T. E. Gough and G. J. Palenik, Jahn–Teller distortions in octahedral copper(II) complexes, J. Am. Chem. Soc., 1969, 91, 1859–1860 CrossRef CAS.
- S. Chaberek Jr and A. E. Martell, Interaction of divalent metal Ions with N-hydroxyethylethylenediaminetriacetic acid, J. Am. Chem. Soc., 1955, 77, 1477–1480 CrossRef.
- L. M. Hellman and M. G. Fried, Electrophoretic Mobility Shift Assay (EMSA) for detecting protein-nucleic acid interactions, Nat. Protoc., 2007, 2, 1849–1861 CrossRef CAS PubMed.
- D. Lane, P. Prentki and M. Chandler, Use of gel retardation to analyze protein-nucleic acid interactions, Microbiol. Rev., 1992, 56, 509–528 CrossRef CAS PubMed.
- R. S. Tuma, M. P. Beaudet, X. Jin, L. J. Jones, C. Y. Cheung, S. Yue and V. L. Singer, Characterization of SYBR gold nucleic acid gel stain: a dye optimized for use with 300-nm ultraviolet transilluminators, Anal. Biochem., 1999, 268, 278–288 CrossRef CAS PubMed.
- B. McGorman, S. Poole, M. Vázquez López and A. Kellett, Analysis of non-canonical three- and four-way DNA junctions, Methods, 2023, 219, 30–38 CrossRef CAS PubMed.
- A. Kellett, Z. Molphy, C. Slator, V. McKee and N. P. Farrell, Molecular methods for assessment of non-covalent metallodrug–DNA interactions, Chem. Soc. Rev., 2019, 48, 971–988 RSC.
- B. McGorman, N. Z. Fantoni, S. O'Carroll, A. Ziemele, A. H. El-Sagheer, T. Brown and A. Kellett, Enzymatic synthesis of chemical nuclease triplex-forming oligonucleotides with gene-silencing applications, Nucleic Acid Res., 2022, 50, 5467–5481 CrossRef CAS PubMed.
- (a) F. A. Taiwo, Mechanism of tiron as scavenger of superoxide ions and free electrons, Spectroscopy, 2008, 22, 491–498 CrossRef CAS; (b) C. Slator, Z. Molphy, V. McKee, C. Long, T. Brown and A. Kellett, Di-copper metallodrugs promote NCI-60 chemotherapy via singlet oxygen and superoxide production with tandem TA/TA and AT/AT oligonucleotide discrimination, Nucleic Acid Res., 2018, 46, 2733–2750 CrossRef PubMed; (c) N. McStay, C. Slator, V. Singh, A. Gibney, F. Westerlund and A. Kelllett, Click and cut: a click chemistry approach to developing oxidative DNA damaging agents, Nucleic Acid Res., 2021, 49, 10289–10308 CrossRef CAS PubMed.
- S. Goldstein, G. Merenyi, A. Russo and A. Samuni, The role of oxoammonium cation in the SOD-mimic activity of cyclic nitroxides, J. Am. Chem. Soc., 2003, 125, 789–795 CrossRef CAS PubMed.
- N. McStay, C. Slator, V. Singh, A. Gibney, F. Westerlund and A. Kellett, Click and Cut: a click chemistry approach to developing oxidative DNA damaging agents, Nucleic Acid Res., 2021, 49, 10289–10308 CrossRef CAS PubMed.
- C. Slator, Z. Molphy, V. McKee, C. Long, T. Brown and A. Kellett, Di-copper metallodrugs promote NCI-60 chemotherapy via singlet oxygen and superoxide production with tandem TA/TA and AT/AT oligonucleotide discrimination, Nucleic Acid Res., 2018, 46, 2733–2750 CrossRef PubMed.
- H. Leonhardt, H. P. Rhan, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink and M. C. Cardoso, Dynamics of DNA replication factories in living cells, J. Cell Biol., 2000, 149, 271–280 CrossRef CAS PubMed.
- R. M. Bringhurst and P. A. Schaffer, Cellular stress rather than stage of the cell cycle enhances the replication and plating efficiencies of herpes simplex virus type 1 ICP0-viruses, J. Virol., 2006, 80, 4528–4537 CrossRef CAS PubMed.
- TUNEL Assay (commercial) Kit detects the DNA fragmentation of apoptotic cells by exploiting the fact that the DNA breaks expose many 3′-hydroxyl ends. These hydroxyl groups can then serve as starting points for terminal deoxynucleotidyl transferase (TdT), which adds deoxyribonucleotides in a template-independent fashion. Addition of the deoxythymidine analog 5-bromo-2′-deoxyuridine 5′-triphosphate (BrdUTP) to the TdT reaction serves to label the break sites. Once incorporated into the DNA, BrdU can be detected by an anti-BrdU antibody using standard immunohistochemical technique.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc03303a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |