Open Access Article
Open Access ArticleImprinting of nanoparticles in thin films: Quo Vadis?
Din
Zelikovich
,
Linoy
Dery
,
Hila
Sagi-Cohen
and
Daniel
Mandler
 *
*
Institute of Chemistry, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel. E-mail: daniel.mandler@mail.huji.ac.il
First published on 2nd August 2023
Abstract
Nanomaterials, and especially nanoparticles, have been introduced to almost any aspect of our lives. This has caused increasing concern as to their toxicity and adverse effects on the environment and human health. The activity of nanoparticles, including their nanotoxicity, is not only a function of the material they are made of but also their size, shape, and surface properties. It is evident that there is an unmet need for simple approaches to the speciation of nanoparticles, namely to monitor and detect them based on their properties. An appealing method for such speciation involves the imprinting of nanoparticles in soft matrices. The principles of imprinting nanoparticles originate from the molecularly imprinted polymer (MIP) approach. This review summarizes the current status of this emerging field, which bridges between the traditional MIP approach and the imprinting of larger entities such as viruses and bacteria. The concepts of nanoparticle imprinting and the requirement of both physical and chemical matching between the nanoparticles and the matrix are discussed and demonstrated.
1. Introduction
This review focuses on a new field of research, which was born as a result of applying the concepts and insights of molecular imprinting to nanochemistry and nanotechnology. The idea that a nanomaterial, which does not have a specific molecular weight but rather a distribution of weights and sizes, can be imprinted similarly to a molecule is not trivial. At present, only a small window has been opened to nanomaterial imprinting and it is already evident that the spectrum of possibilities and applications is wider than expected. How can nanomaterials be imprinted, removed, and detected? What can be expected from the imprinting of nanomaterials? How selective and sensitive can these systems be? What are the foreseen applications of nanomaterial imprinting and what do we learn from the current research status in this field? We will try to answer some of these and other questions that stem from the activity in nanomaterial imprinting. We will begin by briefly reviewing the concept of molecular imprinting and then switch to the imprinting of large entities. Most of the review will describe the concepts and systems where nanomaterials, mostly nanoparticles, have been designed and studied.1.1 Molecular imprinting
Imprinting in materials, refers to the process of printing the shape of an object, i.e., template, in a soft matrix, which creates a cavity with the shape and the outer contour of the imprinted object. Early work in imprinting involved mostly inorganic materials such as silica as the matrix in which organic molecules were imprinted.1 Pauling has introduced the concept of imprinting assuming configurations complementary to surface regions of the antigen to the antibody.2 In the 1950s' molecular imprinting has been applied for the separation of organic molecules, including enantiomers. Yet, the richness of organic chemistry in the design of the matrix, as well as the term “molecular imprinting” has been introduced in the 1970s' by Wulff trying to mimic the action of enzymes.3Molecular imprinting is associated with the imprinting of individual molecules in usually a 3D network that is formed upon polymerization, and therefore, was termed “molecularly imprinted polymers” (MIP). The MIP concept has developed rapidly reaching more than 1000 publications/per year since 2019 (Fig. 1). Excellent reviews and books appeared over the years covering almost any aspect of molecular imprinting and MIPs.4–10
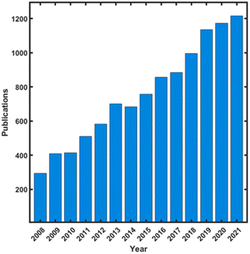 | ||
| Fig. 1 MIP publications over the years (from “Web of Science”, by searching: “molecul* imprint* polym*”). | ||
The vast majority of studies have focused on the imprinting of relatively small organic molecules, i.e., MW < 500, where the primary objective has usually been to form a synthetic receptor that selectively recognizes the imprinted template. Two principal approaches have been applied in the imprinting process; the first involved the formation of covalent hydrolyzable bonds between the template and the matrix, whereas the second approach comprised supramolecular interactions, such as van der Waals and π–π interactions as well as hydrogen bonding. Both approaches can be considered self-assembly where the monomers are arranged around the template as a result of the covalent or supramolecular interactions between the monomers and the template, a process that is followed by polymerization to form either thin films or monoliths.
This “lock and key” approach mimics the activity of natural biological components such as enzymes, antibodies, and aptamers. As such, it demands both thermodynamic and kinetic requirements for proper functionalization. On one hand, the association between the template and its complementary void must be strong, as it affects significantly the selectivity and sensitivity of the system. On the other hand, the ingress and egress of the template to the void must be facile to enable its removal from the cavity. In this respect, it should be mentioned that the major obstacle to the wider application of MIPs is usually the difficulty in extracting the template from the matrix. The extracting methods that are commonly used in MIPs are either based on solvent extraction or physical perturbation, such as microwave and sonication. The removal of the template is also a major concern in the imprinting of nanomaterials, vide infra.
Traditionally, MIPs are formed by bulk polymerization carried out in a solution that consists of the template, the monomer, the cross-linker, and the initiator. The solvent plays a major role and should have a porogenic nature, that is, responsible not only for dissolving properly all the agents during polymerization but also for the development of the porous structure of the MIP. Over the years, a few approaches have been developed for the formation of MIPs.4 These primarily include the conventional bulk polymerization (mentioned above); the polymerization of microemulsions with well-defined initial microstructure, which creates a homogeneous pore distribution and binding sites for analyte recognition;11,12 phase inversion where the MIP is precipitated upon adding an incompatible solvent or by evaporation of the porogen from the solution consisting of the template, and solid-phase synthesis, which involves immobilization of the template on a solid support before carrying out the polymerization,13–15 and surface imprinting polymerization (SIP) through soft lithography or other methods.16,17 The last approach, i.e., SIP, will be discussed in detail later as it is also the basis of more recent large entity imprinting.
Finally, optimizing the imprinting systems is not trivial as it is a multi-parameter challenge, which comprises the proper selection of the functional monomers, cross-linkers, and solvents, and choosing the optimum ratio between the monomers and the template. Therefore, molecular modeling using mostly density functional theory and molecular dynamics methods18–20 are typically used to calculate the binding energy of the host polymer to the template and analyze quite efficiently the matrix–template interactions. These calculations have led to improving the selectivity of MIPs while shortening the time for optimizing their properties.
1.2 Applications
As mentioned above, molecular imprinting has been mostly applied for sensing and separation, although other applications such as in drug delivery can be found. Sensing is by far dominating the application of molecularly imprinting reaching more than 5000 reports. There is no intention to cover this field, which has been reviewed very often and from different aspects.4,5,21 The high selectivity that can be, in principle, achieved by MIPs has made this one of the leading approaches for assembling sensors for small inorganic, organic, and bio-molecules in a wide variety of areas. For example, gas sensors for VOC, explosives,22 and odors have been described,23,24 whereas sensors for the liquid phase have targeted amino acids,25 pesticides,26 pathogens, food adulterants,27 explosives,28,29 environmental pollutants,30 drugs,31 and biomarkers.32 The detection of the template is usually accomplished either electrochemically (by voltammetry, amperometry, potentiometry, conductometry, or via impedance spectroscopy) or optically (by absorption, fluorescence, and surface plasmon resonance) in the UV-visible region. Yet, many substances are electrochemically inactive and absorb only at the far UV and therefore other approaches, such as measuring mass changes by the quartz crystal microbalance33 can also be found. To make these MIP sensors reusable and at the same time increase their sensitivity, imprinting of thin films and high surface areas has been targeted. This has promoted different approaches such as forming nanomaterials and in particular, nanoparticles where the templates were imprinted,34–36 as well as a generic approach of surface imprinting (SIP).17,37 Such an approach of making the recognition layer very thin is also crucial, especially in electrochemical sensors, where electron transfer should be facilitated between the template and the electrode. A recognition layer that is of the order of the size of the template would be ideal and will assure the facile removal of the template and enable to reuse the sensor. This will be discussed in detail as it is of utmost importance in the imprinting and detection of nanoparticles.1.3 Limitations and challenges
Molecular imprinting is a mature well-developed approach and nevertheless has not been very widely commercialized. In spite of the large number of reports covering almost any type of species including small molecules, macromolecules, viruses, and bacteria, molecular imprinting still faces some challenges.38–40 Large molecules such as proteins have rich surface chemistry and therefore require a careful design of the recognition interface and the introduction of additional precursors and the formation of heterogeneous binding sites. The removal of the template still remains a challenge and obstacle in many MIP and SIP systems. A variety of approaches has been developed such as micro liquid–liquid extraction, for improving the process, yet, a rigid matrix where the analyte is well buried inside makes the efficient removal an almost impossible task. Increasing the matrix flexibility by mimicking the natural approach in enzymes and antigens seems also an important unresolved challenge. This enables not only facile kinetics of the ingress and egress of the substrate to the recognition site but also allows water or solvent molecules to maintain their interaction with the analyte, which otherwise requires additional energy. The following sections will deal with the imprinting of large entities and specifically focus on the imprinting of engineered nanomaterials. This is closely related and based on the traditional molecular imprinting methodology and principles.2. Large entities imprinting
2.1 Imprinted objects
The success of small molecule imprinting encourages researchers to expand the field of imprinting to more complex and challenging templates, e.g., proteins,41–44 bacteria,45–53 whole cells,54–57 and NPs.58,59 The common laboratory techniques to analyze biomolecules span from flow cytometry60 to polymerase chain reaction (PCR),61 and enzyme-linked immunosorbent assay (ELISA).62 However, most if not all of these techniques are complex, costly, and require skilled operators. Although these methods achieve the desired detection limits with high specificity, they are complicated for the rapid detection required in various applications, such as medicine and security. There are currently no adequate solutions that allow the selective and sensitive binding to specific microorganisms, that are fast and highly selective, cheap to implement, and able to be conceptualized for a wide range of biologically relevant targets.The detection and characterization of NPs require different approaches and techniques than those used for molecular species. Among these are UV-vis and IR spectroscopy, microscopy (SEM and TEM), light scattering (dynamic and static), centrifugation, electrophoresis, and others.63 A few of these techniques aim at single-particle detection, whereas most are used for particle statistic analysis. Yet, most of these methods are costly and cannot be applied on-site. Moreover, very few provide chemical information about the surface functionalities of the examined NPs. Hence, simple, miniature, and inexpensive in vitro sensors for the detection of NPs are still in their infancy.
The motivation to imprint the above mentioned biological entities or NPs has been primarily for the development of a low-cost and fast platform for different applications such as separation, sensing, etc64–68. However, the urge for imprinting with higher dimensions poses difficulties in the extraction and rebinding steps, as a result of the relatively lower diffusion of the large object as compared with molecules. Therefore, other methods have been developed and particularly the so-called surface imprinting polymers (SIP),17,21,37,69 aiming to improve the imprinting of a wide range of sizes and templates (Fig. 2). Apart from their large size and limited diffusion across the imprinted polymer, these analytes are affected by different conditions such as solvents, monomers, and temperature. Hence, the above-mentioned limitations in the imprinting of large entities led to the development of new imprinting strategies and approaches, which will be described in detail below categorized by the type of the imprinted objects.
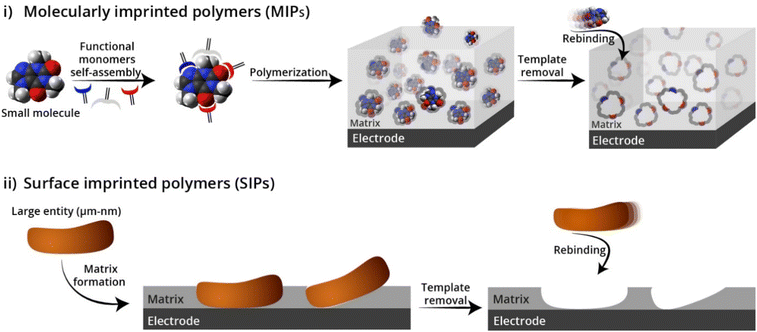 | ||
| Fig. 2 The approaches used for imprinting: (i) molecularly imprinted polymers (MIPs), (ii) surface imprinted polymers (SIPs). Reproduced from ref. 58 with permission from Elsevier, Copyright 2022. | ||
The so-called bacterial imprinting polymer (BIP) and virus imprinted polymers (VIP) are powerful and promising techniques for the selective identification of large bio-entities.79–82 These methods have been developed based on the MIP approach and are mostly applied similarly to surface imprinted polymers (SIPs), that is by imprinting into a thin film deposited on top of a supporting substrate. The intrinsic properties of the bacteria and viruses as well as their membrane topography and chemical properties are the main strategies for developing BIPs and VIPs. Because of the large size, volume, and complexity of the membrane of the bio-template, the chemical recognition comprises mostly the multiple non-covalent interactions between the cell surface and the imprinted layer. For example, proteins are large biomolecules composed of a chain of amino acids. These macromolecules have vast and crucial roles in the structure, function, and regulation of living organisms.83 Proteins unlike small molecules have complicated structures that consist of primary (amino acids sequence), secondary (α helix and the β pleated sheet), tertiary (overall three-dimensional structure), and quaternary (multiple polypeptide chains) structures.84 This complex structure poses an even larger obstacle for the imprinting of proteins by traditional imprinting methods. Thus, the use of organic solvents and different monomers that are frequently employed for the imprinting process is limited, as the three-dimensional protein structure, and conformation depend on the different chemical interactions within the protein, such as hydrogen bonding, dipole–dipole interactions, London dispersion forces, and hydrophobic interactions. The aquatic environment is very important for maintaining these non-covalent chemical bonds that are responsible for protein folding and activity. Moreover, as these bioentities are characterized by a plethora of functional groups, e.g., hydroxyl, carboxyl, and amide, it is easier to bind and dissociate them through non-covalent interactions.
The large-sized microorganisms, which affect the diffusion coefficient, are normally lower than small molecules, which slows down the ingress and egress in and out of the matrix. This limitation can result in low desorption efficiency and poor mass transport. Different imprinting strategies have been developed to enhance the accessibility of the bio objects to the imprinted cavities. Such strategies involve mostly SIP approaches such as nanoscale imprinting,85,86 surface imprinting,85 and soft-gel imprinting.87,88 These imprinting strategies involve creating multiple recognition sites on top of the films and by doing so, prevent the removal problem and avoid slow binding kinetics and low binding capacity. Hence, BIPs and VIPs have the potential of being biosensors with a wide platform of applications.
The dissemination of bioentities detection by the principles of the MIP approach faces three critical challenges that upon resolving, can pave the way to creating effective biosensors. (1) Selecting and controlling the matrix formation for achieving physical recognition, e.g., avoid burying the microorganisms during the imprinting process. (2) Introducing specific functionalities to the matrix, which improves the chemical recognition. (3) Obtaining the optimal interaction between the imprinted cavities and the targetted analyte. A recently published work by Rezai et al. addresses these three challenges. Specifically, they created cell-imprinted polymers (CIPs) on stainless steel microwires through chemical bonding between the functional monomers on the cell and the surface functional groups, and demonstrated that the interaction between the analyte and the surface is crucial for the rebinding capacity.89
In the last decade, few interesting reviews have been published and focused on the recent development in the imprinting of large bioentities. Most of them highlighted new applications and summarized the recent imprinting strategies that have developed in recent times.41,51,54,90–94
To conclude, biomacromolecules as a template for the MIP approach remain challenging due to their complex structure, large size, and environmental instability. The novel bioentities imprinting methods and the improvement of existing techniques for MIP have introduced new applications in separation and purification, biomarker detection, bioimaging, and therapy. Despite the great progress in this field, there are still serval challenges that require further research and development. These include the recognition mechanism between the template and the cavities or the matrix, the synthesis conditions, which must be optimized according to the protein structure, and eventually, the development of real-life applications.
2.2 Imprinting approaches
Proteins, viruses, and bacteria have mostly been imprinted using the SIP approach. The latter can be divided into two; the direct and the indirect methods. Generally, the principle in both methods involves the creation of multiple recognition sites forming non-covalent interactions with favorable binding kinetics.952.2.1.1 Microcontact/stamp imprinting/soft lithography. Microcontact is one of the most common whole-object imprinting processes due to its simplicity and convenience. It enables the formation of a shape recognition site for relatively large templates on surfaces in four main steps. (1) Adsorption of the template onto a solid support substrate. (2) Deposition of a thin layer as a matrix (prepolymer) on top of a transducer such as an electrode. (3) Stamping the functional supporting substrate with the transducer. (4) Removing the stamp with the analyte, leaving an imprint of the template on the transducer. The crucial point of this process is to optimize the viscosity of the prepolymer for generating well-imprinted cavities. Therefore, this technique is unsuitable for some fragile and sensitive templates and is mostly used for small molecules to whole cells, e.g., viruses and bacteria (Fig. 3).45,96–103
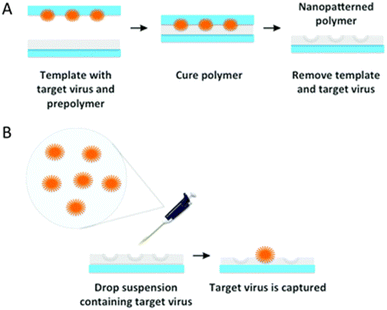 | ||
| Fig. 3 Schematic illustration of (A) VIPs via (B) stamp imprinting. Reproduced from ref. 103 with permission from the Royal Society of Chemistry, Copyright 2015. | ||
2.2.1.2 Drop casting/film imprinting. Compared to the microcontact method, this method can be used for imprinting fragile microorganisms and templates. In this method, the template entities are embedded by drop casting onto the functional surface, immediately after the prepolymer is spin-coated.104,105
2.2.1.3 Sacrificial layers method. The sacrificial layer is used mainly to prevent a reaction between the matrix monomers and the template. This method takes the advantage of surface imprinting polymer, which allows easy removal of the bio-nanoimprinting template from the polymeric matrix. In this method, a sacrificial layer is covalently bound on top of the matrix layer between the sample and the matrix and serves as a transient substrate during the imprinting steps. This method requires complicated preparation and operation.106–108
2.2.1.4 Pickering emulsion interfacial imprinting. Pickering emulsion is a method based on oil–water or water–oil emulsion polymerization. The basic principle is the partitioning of solid particles between two immiscible liquids. This technique involves three main steps. (1) The template is assembled with the prepolymer. (2) The prepolymer and the template are used as stabilizers to form an emulsion of the cross-linked monomer. (3) After imprinting, the polymer beads can be used to capture and detect bacteria59,109–111 (Fig. 4).
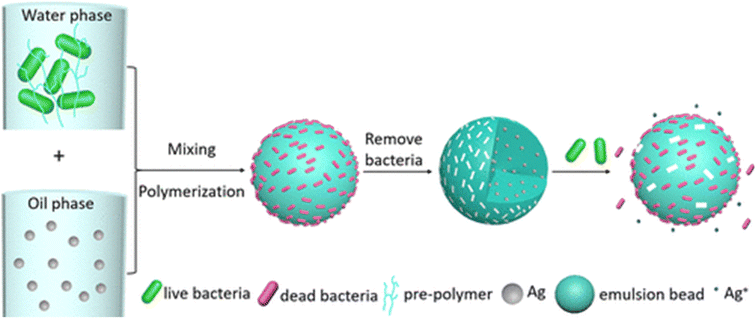 | ||
| Fig. 4 Illustration of surface imprinted of bacteria and Ag NPs via Pickering emulsion polymerization. Reprinting (adapted) with permission from ref. 59. Copyright 2021, American Chemical Society. | ||
2.2.1.5 Electropolymerization. This is a well-established whole-cell imprinting process due to its simplicity, fast preparation, and ability to create a SIP layer with good adhesion to the electrode surface. In this process, the template is entrapped in a matrix via the electropolymerization of electroactive monomers. Hence, a SIP layer forms on top of the electrode surface, where the template is surrounded by a functional matrix (Fig. 5). A major advantage of electropolymerization is the fine control of the SIP thickness by the electropolymerization conditions, such as potential and scan rate. To allow easy removal of the analyte and to form well-defined cavities on top of the electrode surface, it is crucial to deposit a polymeric matrix that does not entirely cover the template.49,112–117
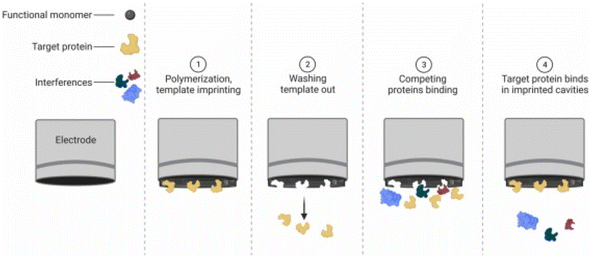 | ||
| Fig. 5 Illustration of BIPs via Electropolymerization. Reproduced from ref. 118 with permission from Elsevier, Copyright 2023. | ||
2.2.1.6 Colloidal imprinting/colloidal antibodies/miniemulsion polymerization. In this method, the whole-cell template is encapsulated by an inorganic shell forming core–shell particles. The most common encapsulation is by the sol–gel process by which mostly a silica-based shell is formed by the polymerization of silane monomers around the microorganism template. The template is removed to form an imprinting site in the silica gel shell, which fits the size and shape of the imprinted template.119–124
2.2.1.7 Molding technique. Usually, molding techniques begin with imprinting the templates on a solid support substrate. Then a functional matrix is a deposit on top of the template layer. In the last step, the solid support is removed and a flexible imprinted layer with high-density imprinted sites was created125–127
The commonly used detection method for pathogenic microorganisms or complex templates in the indirect approach is known as artificial template imprinting. In this method, the natural template is imprinted on the top of the soft polymer matrix. Removal of the template leaves well-defined holes that are filled with a synthetic polymer. That artificial template, i.e., the synthetic polymer, can be used to create cavities for the detection of microorganisms, which fit the initially imprinted template. The preparation procedure for this approach can be cumbersome and usually requires a long time, nevertheless, the recognition can be used for real microorganisms.130–132
3. Nanomaterial imprinting
This is the focus of the entire review, and therefore, will be detailed. The concept, principles of imprinting of nanomaterials, and current insights will be explained. We will end with possible applications and implications of this field to other scientific areas.3.1 The concept
In principle, once molecular imprinting has been extended to larger molecules and even nanometer and micrometer size objects such as bacteria and viruses, vide supra, it should also apply to nanomaterials. Yet, it should be realized that nanomaterials, such as nanoparticles or nanorods, are not molecular materials, namely, they are an assembly of distributed molecular weights, shapes, and properties. Even carefully synthesized gold nanoparticles (NPs) vary substantially in their number of gold atoms and therefore, the meaning of molecular imprinting loses its fundamental basis. This immediately raises issues and doubts as to the ability of imprinting and reuptaking a collection of materials and how well should be the matching between the imprinted template and the cavity. Surprisingly and based on numerous studies, vide infra, imprinting of nanomaterial works very well with high specificity that originates from their size, structure, and surface chemistry. The term NAIM, which refers to nanoparticle-imprinted matrices was introduced recently by Mandler and his coworkers.133,134The approach has been so far similar to molecular imprinting; however, the process has always involved the formation of a thin film, where the nanomaterials were imprinted (Fig. 6). Various methods have been developed and will be discussed below for the formation of the matrix and controlling its thickness. The latter was found to be of utmost importance and it impacts both the ability to remove the embedded nanomaterials as well as to reuptake them. Due to the ease of preparation and their high symmetry, NPs have always been used as templates. Furthermore, the removal of the nanomaterials is also a crucial step, and therefore, applying stable, yet, oxidizable NPs made of either gold or silver has usually been preferred. This enabled their electrochemical dissolution, which was also used as a means of determining the amount and percentage of reuptaken NPs based on the oxidation charge. Evidently, for the detection of non-conductive nanomaterials, other methods, such as capacitive and change of mass, will have to be developed and tested.
 | ||
| Fig. 6 Schematics of the NAIM approach: adsorption of the NPs, forming the matrix in the non-occupied areas, removing the NPs, and finally reuptaking the NPs. | ||
Imprinting of NPs has some interesting differences and also advantages over molecular imprinting, which stem from their nanometric size, rigidity, and uniform surface chemistry. For example, the nanometric size enables imaging of individual imprinted cavities and particles, while the rigidity makes it possible to obtain cavities with a distinct shape. Moreover, taking into account that the interactions between the nanomaterials and the cavity are based on the surface chemistry of the particles allows imprinting with one nanomaterial for the recognition of another one, providing that both possess similar surface functionalities. Hence, successful imprinting requires both physical as well as chemical matching between the template and the imprinted matrix.
3.2 Imprinting approaches
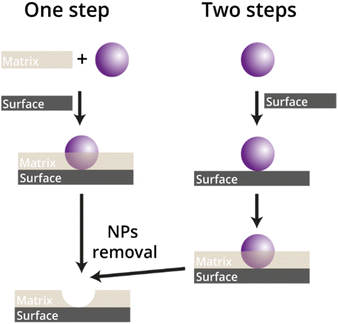
The formation of a NAIM system comprises imprinting the nanomaterials followed by their removal to form nanometric voids. The latter is mainly used for the selective reuptake of the imprinted NPs. Two main strategies for imprinting NPs in functional matrices (Fig. 7) have been demonstrated.In the so-called one-step approach the NPs and the matrix are co-deposited simultaneously onto the substrate to form the NAIM. This concept has primarily used the Langmuir Blodgett (LB) technique, whereby monolayers of the polymer constituting the matrix are deposited together with NPs already embedded inside the polymer. This method enables controlling precisely the thickness of the film by the number of deposited monolayers as well as optimizing the NPs–matrix interactions due to mixing the components before deposition. On the other hand, since each deposition contains both the matrix and the NPs, the upper monolayer might cover the NPs of the lower layer and inhibit their extraction thus diminishing the recognition ability. In the second, two-step approach, the NPs are first adsorbed on the surface followed by filling the unoccupied areas with the matrix through electropolymerization or dip- or spin-coating. The main advantage of the two-step approach is the excellent control of the density of the NPs adsorbed on the surface and the formation of a single NP layer. Yet, special attention should be paid to avoid covering the NPs with the matrix, which will prevent their partial removal. Following is an in-depth description of both approaches.
Three main strategies based on the one-step approach, called nanoparticle imprinted polymers have been demonstrated. The first and simple method for polymer-embedded NPs involved blending the NPs135 with polymers or the reduction of metal salts in a molten polymer.136 However, these approaches could not form interactions between NPs and the matrix such as supramolecular interaction or a hydrolyzable chemical bond, which is necessary for specific recognition. Moreover, this straightforward method is hampered by several difficulties including the characterization of the metallic ions and the NPs in the polymer, their solubility and dispersibility, and the aggregation of the NPs, which can lead to their uneven dispersion in the polymer. To overcome these obstacles, a second method was introduced that involved the incorporation of ligand shells in the NPs as cross-linkers in the bulk polymers. For example, Lev et al. showed that AuNPs can be stabilized and dispersed by organically modified silica sol without additional external stabilizing agents. Sol–gel-based matrices with different thicknesses where the NPs were embedded, were formed by dip or spin coating.137 This approach has further been investigated by Kulkarni et al. who synthesized AuNPs in the presence of poly bis(methacryloyl hydroxyethyl)disulfide (DSDMA).138 Dissolution of the AuNPs by methyl iodide resulted in the formation of nanocavities with the size of the template. Yet, the matrix was not used for recognition of the template. Chechik et al.139 used the same approach, in which the NPs were stabilized by the monomers of the matrix for the reuptake of NPs. More specifically, AuNPs were stabilized by an acrylate-based ligand that was polymerized (by the added initiator) and cross-linked to form the matrix. They concluded that a porogen was essential to create a solvent-permeable and highly porous matrix required to dissolve the NPs. Moreover, to prepare a rigid structure of cavities with the shape of the template, large quantities of cross-linking agents were needed. Eventually, their system showed selective adsorption of small AuNPs. In a more recent study, Schwob et al.140 described the selective recognition of quantum dots differing in their size and structure by quantum dot imprinted polymers (QD@IPs) (Fig. 8). The selective system was prepared by photopolymerization of a carboxyl-functionalized QD and methacrylic acid as a monomer upon adding a cross-linker and a photoinitiator. The matrix and the functionalized QDs interacted through strong hydrogen bonding. The addition of acetic acid caused the cleavage of these interactions and the removal of the QDs. The recognition of their system was examined by QDs having different sizes and shells and studied through photoluminescence. They showed highly efficient reuptake of the initially imprinted QDs, which evidenced the attractive interactions between the target analyte and the matrix.
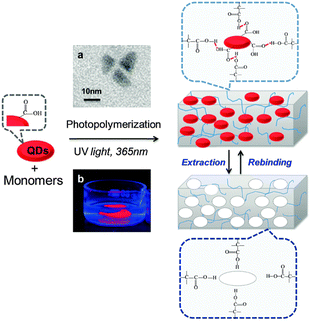 | ||
| Fig. 8 Schematic illustration of the elaboration of bulk QD-imprinted polymers QD@IPs. (a) TEM image of the QD templates and (b) photography under UV light (at 254 nm) of bulk as-synthesized QD@IPs immersed in distilled water. Reproduced from ref. 140 with permission from the Royal Society of Chemistry, Copyright 2015. | ||
There are serval drawbacks to this method, which include complicated preparation, additions of a porogen, difficult identification of the cavities and the NPs inside the matrix by microscopy tools, and the difficulty in the removal of the template from the matrix. These challenges led to the development of the third strategy in which the NPs and the matrix are incorporated into monolayers, which allows their simple removal and reuptake.
More specifically, the Langmuir–Blodgett (LB) technique has been used, whereby NPs were embedded into monolayers based on amphiphilic polymers formed at the water–air interface and deposited onto a solid electrode. The LB method offers several advantages such as controlling very precisely the thickness of the matrix and the NPs by deposition of multilayers reproducibly. Moreover, co-spreading of the NPs and the matrix followed by their compression enables to increase of the NPs surface density, and at the same time, forces them to interact with the matrix. On the other hand, since each deposition contains both the matrix and the NPs, the upper monolayer might cover the NPs of the lower layer, thus, inhibiting the removal reuptake of all the NPs.
The LB approach has been applied to form patterns,141,142 nanostructured films,143,144 and 2D nanocomposites. The Langmuir monolayer is usually made of either amphiphilic molecules or polymers that can be used to attract and accommodate nanomaterials. The latter can be either co-spread with the LB monolayer or dissolved in the aqueous subphase. For example, Kalinina et al.145 co-deposited gold NPs (AuNPs) stabilized with citrate (AuNPs-cit) and a mixture of stearic acid and octadecyl amine. They found that the addition of the AuNPs-cit induced a rapid condensation of the mixed surfactant as well as the formation of two-dimensional gel-like network of the AuNPs-cit. The adsorption of AuNPs-cit within the mixed monolayer led to an increase in the interactions between oppositely charged surfactants, i.e., stearic acid and octadecyl amine, giving a “soap” of mixed fatty salts. The rearrangement of the monolayer resulted in the aggregation of the NPs into colloidal 2D “soap”-gels, which represented arrested colloidal phases within a nonadsorbing organic medium.
Huang et al. controlled the density of the NPs in a LB film by the transfer speed and the NPs concentration in the Langmuir film.146 Hence, the combination of an organic layer and NPs at the air–water interface was used for the fabrication of surface-enhanced Raman spectroscopy (SERS) surfaces. Adding a low-dielectric solvent caused the movement of metallic NPs from the aqueous subphase to the water/oil interface147 and led to the homogeneity of the film surface which provided 10 times stronger SERS signal as compared with the free surface. The application of the LB technique as a sensing tool was examined for the selective recognition of different enzymes such as glucose-oxidase and urease by Harrison and his coworkers.148
To the best of our knowledge, the first study where the LB method was used for the imprinting of NPs was reported by Kraus-Ophir et al.133 Specifically, AuNPs-cit were dispersed in the aqueous subphase while polyaniline (PANI) as a matrix layer was spread at the water–air interface. Under mildly acidic pH, the AuNPs were negatively charged and attracted by the positively charged PANI. In the next step, indium-doped tin oxide (ITO) as solid support was vertically withdrawn across the air–water interface causing the deposition of AuNPs-cit/PANI. Controlling the number of layers was of paramount importance and beneficial for the formation of the first NAIM system where the AuNPs were embedded inside, yet protruding through the PANI layers (Fig. 9). The AuNPs were anodically dissolved and the size exclusion of AuNPs by the NAIM system was studied. The system showed remarkable selectivity, and the nano-cavities formed in the PANI could be nicely imaged by electron microscopy.
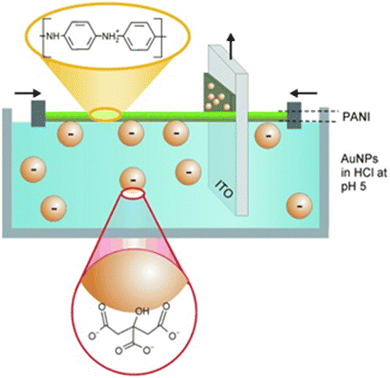 | ||
| Fig. 9 Schematic representation of the LB approach for assembling a matrix with embedded nanoparticles. Reproduced from ref. 133 with permission from Elsevier, Copyright 2014. | ||
It is worth mentioning that some of the AuNPs remained unoxidized due to the covering and isolation of the PANI under the experimental condition, which presumably hindered the chloride access necessary for dissolving the AuNPs. The recognition was determined by studying the ratio, i.e., reuptake percentage, of the charge associated with the reuptake process with that of the initial removal of the template. It was found that the oxidation charge increased with the number of the deposited layer because each layer contained more NPs, and thus, more cavities were formed that could be used for the reuptake. Moreover, this NAIM system was found to be optimal for size-selective recognition and depended directly on the thickness of the matrix layer or the depth of the cavities. In addition, the authors emphasized the importance of the supramolecular interactions between the NPs and the matrix, e.g., hydrogen bonding, after the reuptake process, which caused the NPs to remain in the cavities.
Another study that was carried out by Mandler et al. uses the LB method for studying the effect of the alkanethiol chain stabilizing the AuNPs on the reuptake selectivity in a NAIM system.149 The system consisted of four consecutive stages (Fig. 10A) similar to the previous one. Initially, the 2 nm AuNPs stabilized with 1-dodecanethiol (AuNPs/C12) were co-spread with cellulose acetate (CA) at the air–water interface from an organic solvent. The next step comprised the compression of the Langmuir film (Fig. 10B), which was followed by the deposition on an ITO surface vertically withdrawn from the subphase. To increase the density of the AuNPs on the ITO and the thickness of the CA matrix (which was ca. 1 nm per layer) this process was repeated several times. The oxidation of the AuNPs and the reuptake process were carried out in a chloride solution. The detection of an oxidation peak approved that the CA layers did not fully cover the AuNPs, which enabled their electrochemical dissolution.
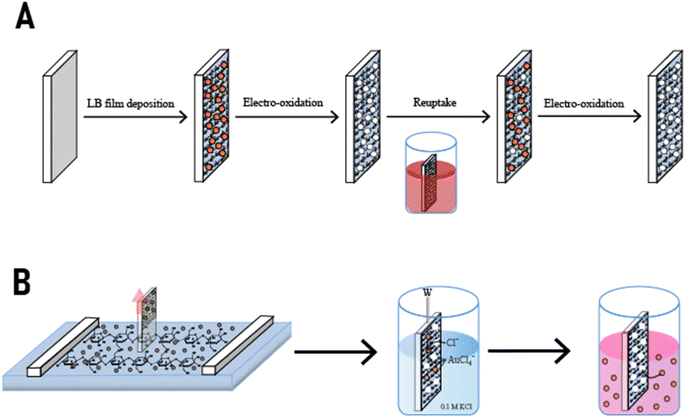 | ||
| Fig. 10 The different steps in the formation of a NAIM system based on different lengths of alkanethiol stabilized AuNPs. Reproduced from ref. 149 with permission from Elsevier, Copyright 2015. | ||
It was reported that the recognition ability of the AuNPs/C12 increased with the number of the CA LB layers. Specifically, beyond two layers, the reuptake capability improved significantly due to the formation of deeper nanocavities, which increased the attractive supramolecular interactions between the AuNPs/C12 and the NAIM matrix. Furthermore, the system exhibited very high selectivity, with high attraction toward AuNPs with the same alkanethiol capping agent as the originally imprinted AuNPs, i.e., AuNPs/C12, but almost no recognition of AuNPs with either longer or shorter chain length of the alkanethiol. The high selectivity indicated the formation of tight wrapping of the AuNPs/C12 by the matrix, and therefore, the nanocavities could not accommodate well AuNPs bearing different alkanethiol capping agents.
To summarize, despite the straightforward imprinting in a single step, the one-step approach suffers from several disadvantages such as the ability to grow the matrix well around the NPs and the lack of ohmic contact between the NPs that are deposited in the repeated deposition cycles and the surface.
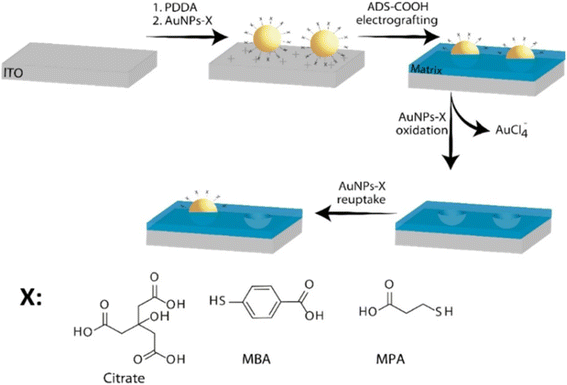 | ||
| Fig. 11 Schematics of the NAIM system: ITO substrate is treated with PDDA followed by adsorption of AuNPs-X where X defines the chemical structure of the different ligand shells that were used. Then, the aryldiazonium-based matrix was electrografted on the electrode to form the NAIM. The AuNPs-X were removed by electrochemical oxidation to form the template, which was used to reuptake the same AuNPs-X. Reprinting (adapted) with permission from ref. 150. Copyright 2021, American Chemical Society. | ||
Thus, the development of the new two-step fabrication process was crucial to the extension of the NAIM concept to different matrices and analytes. In this approach, the NPs are attached to the surface using primarily electrostatic interaction. For example, different surfaces, e.g., ITO, silicon, and Au, were functionalized by adsorbing a polyelectrolyte, such as polydiallyldimethylammonium chloride (PDDA) and polyethyleneimine (PEI) followed by the adsorption of negatively charged NPs stabilized with citrate or other functionalized carboxylic acids.150–152 A clear correlation between the polymer concentration and the density of the adsorbed AuNPs stabilized with citrate was demonstrated for different PEI concentrations.153 The physical deposition of NPs on the surface was also demonstrated by spin-coating. This method enabled to deposit NPs without functionalization of the surface. However, spin-coating is limited to flat surfaces with limited shapes and the NPs tend to aggregate on the surface.154
Following NPs deposition, the matrix is formed around the NPs by a controlled process such as electropolymerization, adsorption, and dip and spin-coating. Controlling the thickness of the matrix was found to be of utmost importance as will be described below. In most systems so far, the formation of the matrix was performed in a solution where the film grew from the solid/liquid interface to the solution. Therefore, electropolymerization was found to be particularly advantageous. Different electropolymerization processes have been used and include aryldiazonium precursors, sol–gel electrodeposition, and phenol oxidation.
Aryldiazonium electropolymerization has extensively been studied150,151 and involves the reduction of an aryldiazonium salt to evolve molecular nitrogen and generate an aryl radical (Fig. 12). The aryldiazonium chemistry offers a wide range of monomers formed by the chemical functionalization of the aryldiazonium salt in the para position. This afforded the formation of aryldiazonium-based matrices with different functionalities on electrode surfaces. Furthermore, this electropolymerization is usually conducted in organic solvents, such as acetonitrile under very mild negative potentials, which is harmless to the attached NPs. The thickness of the layer was controlled very precisely by the number of potential cycles and the scan rate.150,151
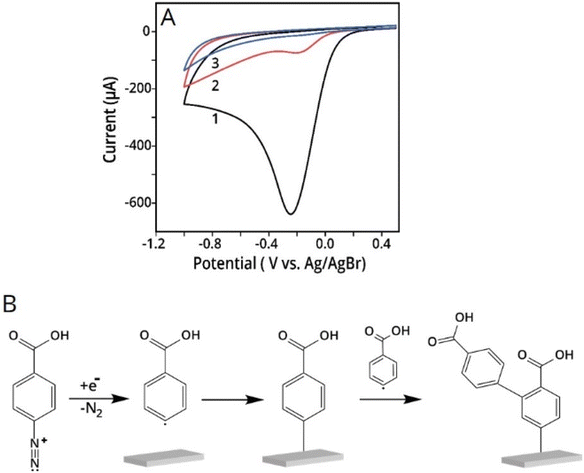 | ||
| Fig. 12 (A) Three repetitive CV scans of conductive electrode covered with AuNPs-cit in aryldiazonium solution. The scan rate was 0.1 V s−1. (B) Radical electrografting mechanism of diazonium precursor on a conductive electrode surface. Reprinting (adapted) with permission from ref. 150. Copyright 2021, American Chemical Society. | ||
Analysis of the matrix by electron microscopy and electrochemistry revealed that the polymer was formed primarily between the NPs although recent results indicate that it can also be partially formed on top of conductive NPs. This allowed the facile removal of the NPs by electrooxidation to form the open nanocavities, which were further used for the reuptake test.58
Mandler's group reported several studies where the aryldiazonium-based matrices were used for the imprinting of NPs. The first study demonstrated the effect of the formation of two different p-aryldiazonium-based NAIM on the reuptake selectivity.151 Specifically, AuNPs-cit were preadsorbed onto a conductive surface followed by electrografting of two different p-aryldiazonium salts (ADS) with different functional groups (hexyl-ether and hexyl-ester). The AuNPs-cit were removed by electrochemical dissolution and the recognition ability of the imprinted diazonium NAIMs was determined by the reuptake of the original NPs. It was demonstrated that a subtle change in the functional group of the p-aryldiazonium building block significantly affected the ability of the NAIMs to recognize the imprinted NPs. Additional studies by Mandler and his co-workers clearly show that the nature of the aryldiazonium-based matrix and its interaction with the imprinted NPs affects the NP recognition.150,155 These NAIM systems take advantage of the aryldiazonium chemistry that enables the covalent binding of a wide range of matrices to a variety of surfaces. The matrix–NP interactions are largely dependent on the surface chemistry of the NPs, the substituted aryldiazonium, and the supramolecular interaction between them. Thus, these two studies further develop the NAIM approach focusing on the ability to fine-tune the NP-matrix interactions through modification of both the matrix precursor and the stabilizing shell of the NP. In both cases, the NAIM systems were prepared by imprinting identical size AuNPs, bearing different carboxylic acid-functionalized thiols as capping agents and an ADS-based carboxylic acid matrix. In all cases, high reuptake percentage of the originally imprinted NPs was observed, indicating excellent chemical recognition of the NPs by the imprinted voids of the matrix. It is demonstrated that a relatively small alteration of the capping agent significantly influences the selectivity (Fig. 13).
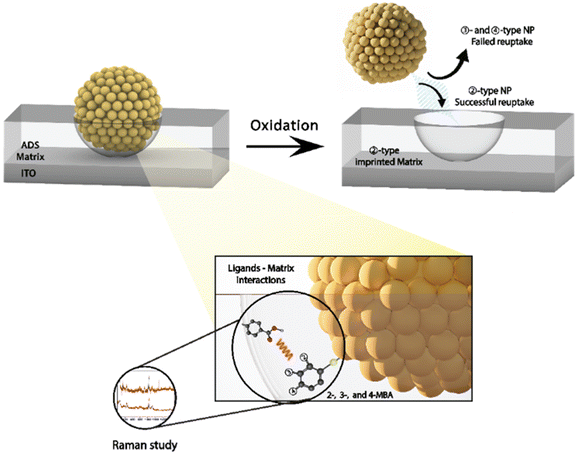 | ||
| Fig. 13 Schematics of the NAIM approach based on different isomers of MBA and schematic illustration of the matrix–ligand interactions. Reprinting (adapted) with permission from ref. 155. Copyright 2023, American Chemical Society. | ||
An alternative method for the formation of a well-controlled thin matrix growing from the electrode/electrolyte interface involves the electrochemical deposition of sol–gel.156,157 This indirect electrodeposition approach was developed by Mandler et al. and is based on enhancing the hydrolysis and condensation processes of the sol–gel precursors by applying negative potentials in protic media. The processes are accelerated under alkaline conditions that are formed near the electrode surface where the reduction of water or alcohol occurs.157–159 The thickness of the sol–gel layer can be controlled by the time and the potential applied.152 Furthermore and due to the availability of numerous sol–gel precursors, it is possible to tailor the physicochemical properties of the matrix, such as its hydrophobicity or porosity. Usually, sol–gel thin films are more permeable to small inorganic and organic species, yet, the electrodeposited thin films are impermeable to NPs. The films have to be post-treated, i.e., drying under controlled conditions, after deposition to avoid cracking.
An example of the formation of NAIM based on electrochemical sol–gel deposition was demonstrated by the authors of this review152 (Fig. 14). Specifically, ITO films were modified with a positively charged polymer, on which the negatively charged AuNPs-cit were adsorbed. This was followed by the electrochemical deposition of a sol–gel matrix with a controlled thickness and various functional groups filling the unoccupied areas of the ITO/AuNPs-cit surface. Electrochemical oxidation dissolved the AuNPs-cit and formed cavities in the sol–gel films, which fit both the size and shape of the AuNPs-cit. Reuptake of these NPs from an aqueous solution was successful using the imprinted films, whereas the non-imprinted films did not reuptake the AuNPs-cit. Furthermore, the thickness of the sol–gel layers as well as the type of silanes that were deposited played an important role in the recognition ability of the NAIM. Hitrik et al. described a different approach to forming a matrix via the self-assembly of oleic acid (OA) or poly(acrylic acid) (PAA).153 Specifically, the authors proved that the selectivity was further improved by thickening the PAA template by self-assembly of an additional 1-hexadecyl amine layer. Furthermore, this NAIM system showed that the voids recognized different core NPs by their surface and geometric characteristics. More precisely, they showed that NPs made of different materials, such as silver and gold, could be reuptaken provided that their size and shell matched those of the template nano-voids.
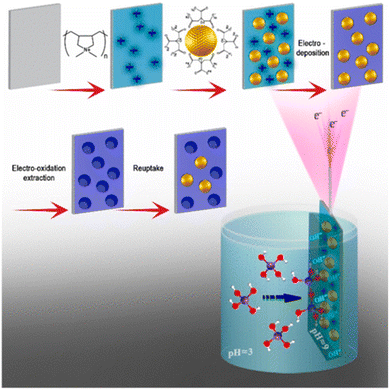 | ||
| Fig. 14 Schematics of the NAIM electrochemical sol–gel deposition system. First, ITO substrate is treated with PDDA followed by adsorption of AuNPs-cit. Then, the desired silane is electrodeposited on the electrode to form the NAIM. Reprinting (adapted) with permission from ref. 152. Copyright 2018, the American Chemical Society. | ||
Another study that was carried out by Wittstock et al. uses electropolymerization of a self-inhibiting polyphenol (PPh) to generate a polymeric matrix.134 The reuptake by this NAIM system (after removing the NPs) was tested by competitive recognition, in which the selectivity of the NAIM was examined in a mixture of two different core metallic NPs, i.e., AuNPs and AgNPs, of different sizes but identical ligand shells. The size selectivity of the films was demonstrated by significantly higher reuptake of the 20 nm AgNPs since they were accommodated in the cavities imprinted by a template of 40 nm diameter, which was not the case for the 50 nm AuNPs. The preference for 20 nm AgNP over 50 nm AuNP greatly exceeded the expected difference in the diffusion of the two size-NPs. More recently, Dery et al.160 exhibited a NAIM approach for the electrochemical detection of non-conductive silica-NPs (Fig. 15). Specifically, the blocking of the nano-cavities was indirectly monitored by recording the redox-activity of hexacyanoferrate(III) inside the nanocavities following exposure to silica-NPs using cyclic voltammetry (CV). The reuptake of negatively charged silica-NPs by polyphenol and poly(3-aminophenol) imprinted matrices was studied. It was shown that the poly(3-aminophenol) matrix displayed improved reuptake as compared with the polyphenol matrix, which was attributed to the difference in the electrostatic attractive forces between the positively charged ammonium functional groups and the negatively-charged silica-NPs.
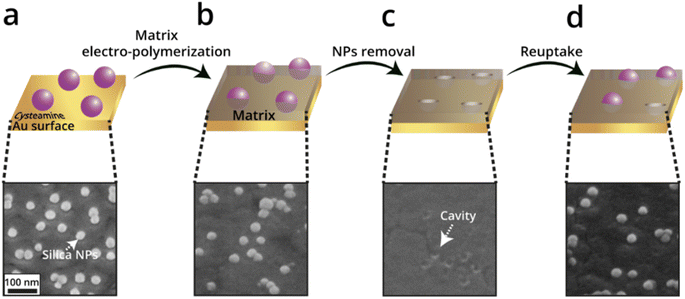 | ||
| Fig. 15 Schematic illustration of the NAIM approach preparation steps (a–d) and the corresponding SEM images of poly(3-aminophenol) matrix as a representative. Reproduced from ref. 160 with permission from Elsevier, Copyright 2023. | ||
Unlike the electrochemical methods that were mentioned above, the two other approaches, i.e., spin-coating and adsorption through SAMs can be used to prepare a thin matrix on non-conductive surfaces. For example, the spin-coating method was used to form an imprinted sol–gel layer in the non-conductive space between two microelectrodes at an interdigitated electrodes array (IDEs) surface as shown in Fig. 16.161 This was achieved by spin-coating of dispersed sol–gel polymer on an IDE surface with deposited 100 nm latex NPs. The latter was also spin-coated under controlled conditions to form a well-distributed mono-particle layer. The thickness of the sol–gel layer was controlled by altering the rotational speed leading to a sol–gel layer with a thickness of ca. 45 nm. The imprinted IDE demonstrated size-selectivity only to silica NPs with dimensions of 100 nm that were similar in size to the nano-cavities.161
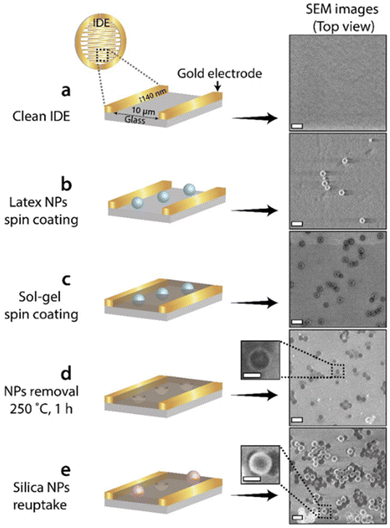 | ||
| Fig. 16 Schematic illustration of the NAIM-IDE-based approach (left) and the corresponding SEM images (right): (a) a bare IDE; (b) following spin-coated with latex-NPs; (c) upon spin-coated with sol–gel; (d) after removal of the latex NPs; (e) following the reuptake of 100 nm silica NPs. Scale bar: 200 nm. Reprinted (adapted) with permission from ref. 161. Copyright 2022, American Chemical Society. | ||
In all cases where the matrix was built around the NPs, it appeared that the layer thickness has a critical role in the reuptake of NPs by the formed nanocavities. This is illustrated in Fig. 17A layer whose thickness is thinner than the NP radius is too shallow to physically capture effectively NPs. On the other hand, a layer whose thickness is too thick, i.e., thicker than the NP radius, makes it almost impossible to either enter or remove the NP into and from the cavity. Hence, the optimal thickness of a matrix so that it will effectively reuptake and release NPs should be close to the radius of the NPs imprinted in it.
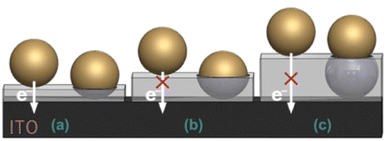 | ||
| Fig. 17 Schematic illustration showing how the layer thickness affects the reuptake process. (a) A very thin layer allows electron transfer of non-specific interactions. (b) The optimal condition where the layer is thick enough to simultaneously prevent electron transfer from non-specific interactions and allow the reuptake of the NPs. (c) Electron transfer of non-specific interactions is prevented, but reuptake of the NPs is impossible as a result of a physical barrier. Reproduced from ref. 151 with permission from Springer, Copyright 2019. | ||
Additionally, the ratio between the signals of the non-specifically adsorbed NPs, i.e., NPs adsorbed on top of the matrix and not in the cavities, and the reuptake NPs is significantly affected by the layer thickness. Specifically, metallic NPs that can be electrochemically oxidized and are non-specifically adsorbed will not contribute to the total current when the layer thickness prevents electron transfer (Fig. 17C). This will increase the signal-to-noise ratio contributed by only NPs accommodating the imprinted nanocavities.
In conclusion, the two-step approach seems to be ideal for imprinting of NPs in soft matrices due to its versatility, ease of preparation and enhanced signal-to-noise ratio. The limitation of this approach stems from the inability to form thick non-conductive matrices by electropolymerization as well as the partial growth of the matrix on top of conductive NPs.
3.3 Applications and perspectives
It is too early to predict the full spectrum of applications where the imprinting of nanomaterials could be disseminated. Yet, it is conceivable that the approaches described above can be beneficial in, at least, sensing as well as separation. The former is straightforward and currently drives the development of this field. The detection of nanoparticles made of different materials and especially carbonaceous particulate in the air is of great interest. To the best of our knowledge, there are nowadays no commercially available sensors for PM0.1 that can differentiate NPs according to their shape, size, and surface properties. The detection of NPs in the air poses substantial challenges as to the method of detection, which will likely involve either monitoring the resistance or capacitance changes. Moreover, more advanced signal analysis approaches including AI will have to be introduced as a means of detecting NPs with a range of sizes such as that found in air pollution.At the same time, the imprinting of NPs has still not been applied to separation as opposed to MIPs. The latter has shown great promise as a versatile and effective tool in chromatography, offering advantages over traditional chromatographic techniques in terms of selectivity, stability, and reproducibility. Nanoparticle imprinting and especially NAIM can similarly be applied for the separation of nanomaterials. Imprinted NPs could be utilized to form micro- and ultra-filtration membranes for the separation and removal of nanoparticles based on their size, shape, and surface properties. The imprinted layer could be formed on a solid porous support where the density of nanopores will readily be controlled by the density of the adsorbed or deposited NPs. Moreover, it can be envisioned that NP imprinting could be used for nanopore DNA sequencing. The size of the pores used for nanopore sequencing typically ranges from 1 to 3 nanometers in diameter, which could be achieved by imprinting of NPs. To accomplish nanopore sequencing, it will be necessary to design free-standing membranes with a known number of nanopores.
Clearly, this area of research, which is in its infancy, requires more effort and additional researchers to advance it and eventually generate also commercial systems such as sensors or membranes. The efforts will presumably be directed toward developing imprinting of a variety of other NPs, detection methods both in liquid and in particular in the air, increasing the reuptake rate and recycling the systems, and improving the analysis of the transduction signals. Yes, we believe that the future of nanoparticle imprinting is promising and that this review will encourage more players to enter this exciting field of research.
Author contributions
Din Zelikovich – writing – original draft, writing – review & editing: Linoy Dery – writing – original draft: Hila Sagi – writing – original draft: Daniel Mandler – conceptualization, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are not conflicts to declare.Acknowledgements
This research was supported by the Israel Science Foundation (research 1953/22).References
- M. V. Polyakov, Adsorption Properties and Structure of Silica Gel, Zhurnal Fizieskoj Khimii, 1931, 2, 799–805 Search PubMed.
- L. Pauling, A Theory of the Structure and Process of Formation of Antibodies, J. Am. Chem. Soc., 1940, 62, 2643–2657 CrossRef CAS.
- G. Wulff and A. Sarhan, Use Of Polymers With Enzyme-Analogous Structures For Resolution Of Racemates, Angew. Chem., Int. Ed., 1972, 11, 341 CAS.
- J. J. BelBruno, Molecularly Imprinted Polymers, Chem. Rev., 2019, 119, 94–119 CrossRef CAS PubMed.
- L. X. Chen, X. Y. Wang, W. H. Lu, X. Q. Wu and J. H. Li, Molecular imprinting: perspectives and applications, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- L. X. Chen, S. F. Xu and J. H. Li, Recent advances in molecular imprinting technology: current status, challenges and highlighted applications, Chem. Soc. Rev., 2011, 40, 2922–2942 RSC.
- K. Haupt and K. Mosbach, Molecularly imprinted polymers and their use in biomimetic sensors, Chem. Rev., 2000, 100, 2495–2504 CrossRef CAS PubMed.
- K. Haupt, P. X. M. Rangel and B. T. S. Bui, Molecularly Imprinted Polymers: Antibody Mimics for Bioimaging and Therapy, Chem. Rev., 2020, 120, 9554–9582 CrossRef CAS PubMed.
- G. Wulff, Molecular Imprinting In Cross-Linked Materials With The Aid Of Molecular Templates – A Way Towards Artificial Antibodies, Angew. Chem., Int. Ed. Engl., 1995, 34, 1812–1832 CrossRef CAS.
- G. Wulff, Enzyme-like catalysis by molecularly imprinted polymers, Chem. Rev., 2002, 102, 1–27 CrossRef CAS PubMed.
- T. V. Nicolescu, W. Meouche, C. Branger, A. Margaillan, A. Sarbu, V. Fruth and D. Donescu, A new microemulsion approach for producing molecularly imprinted polymers with selective recognition cavities for gallic acid, Polym. Int., 2013, 62, 949–956 CrossRef CAS.
- H. Liu, G. Fang and S. Wang, Molecularly imprinted optosensing material based on hydrophobic CdSe quantum dots via a reverse microemulsion for specific recognition of ractopamine, Biosens. Bioelectron., 2014, 55, 127–132 CrossRef CAS PubMed.
- A. Poma, A. Guerreiro, M. J. Whitcombe, E. V. Piletska, A. P. F. Turner and S. A. Piletsky, Solid-Phase Synthesis of Molecularly Imprinted Polymer Nanoparticles with a Reusable Template-“Plastic Antibodies”, Adv. Funct. Mater., 2013, 23, 2821–2827 CrossRef CAS PubMed.
- H. Y. Wang, T. Kobayashi and N. Fujii, Molecular imprint membranes prepared by the phase inversion precipitation technique, Langmuir, 1996, 12, 4850–4856 CrossRef CAS.
- M. Ramamoorthy and M. Ulbricht, Molecular imprinting of cellulose acetate-sulfonated polysulfone blend membranes for Rhodamine B by phase inversion technique, J. Membr. Sci., 2003, 217, 207–214 CrossRef CAS.
- L. Dery, D. Zelikovich and D. Mandler, Electrochemistry of molecular imprinting of large entities, Curr. Opin. Electrochem., 2022, 34, 8 Search PubMed.
- C. Y. Dong, H. X. Shi, Y. R. Han, Y. Y. Yang, R. X. Wang and J. Y. Men, Molecularly imprinted polymers by the surface imprinting technique, Eur. Polym. J., 2021, 145, 25 CrossRef.
- S. Srebnik, Theoretical investigation of the imprinting efficiency of molecularly imprinted polymers, Chem. Mat., 2004, 16, 883–888 CrossRef CAS.
- I. A. Nicholls, H. S. Andersson, C. Charlton, H. Henschel, B. C. G. Karlsson, J. G. Karlsson, J. O'Mahony, A. M. Rosengren, K. J. Rosengren and S. Wikman, Theoretical and computational strategies for rational molecularly imprinted polymer design, Biosens. Bioelectron., 2009, 25, 543–552 CrossRef CAS PubMed.
- T. Cowen, K. Karim and S. Piletsky, Computational approaches in the design of synthetic receptors – A review, Anal. Chim. Acta, 2016, 936, 62–74 CrossRef CAS PubMed.
- K. Eersels, P. Lieberzeit and P. Wagner, A Review on Synthetic Receptors for Bioparticle Detection Created by Surface-Imprinting Techniques-From Principles to Applications, ACS Sens., 2016, 1, 1171–1187 CrossRef CAS.
- G. Bunte, J. Hurttlen, H. Pontius, K. Hartlieb and H. Krause, Gas phase detection of explosives such as 2,4,6-trinitrotoluene by molecularly imprinted polymers, Anal. Chim. Acta, 2007, 591, 49–56 CrossRef CAS PubMed.
- Y. M. Zhang, J. Zhang and Q. J. Liu, Gas Sensors Based on Molecular Imprinting Technology, Sensors, 2017, 17, 14 CrossRef.
- S. Gaggiotti, F. Della Pelle, M. Mascini, A. Cichelli and D. Compagnone, Peptides, DNA and MIPs in Gas Sensing. From the Realization of the Sensors to Sample Analysis, Sensors, 2020, 20, 28 CrossRef PubMed.
- M. Kempe and K. Mosbach, Separation Of Amino-Acids, Peptides And Proteins On Molecularly Imprinted Stationary Phases, J. Chromatogr. A, 1995, 691, 317–323 CrossRef CAS PubMed.
- A. L. Jenkins, R. Yin and J. L. Jensen, Molecularly imprinted polymer sensors for pesticide and insecticide detection in water, Analyst, 2001, 126, 798–802 RSC.
- C. C. Villa, L. T. Sanchez, G. A. Valencia, S. Ahmed and T. J. Gutierrez, Molecularly imprinted polymers for food applications: A review, Trends Food Sci. Technol., 2021, 111, 642–669 CrossRef CAS.
- W. Lu, M. Xue, Z. B. Xu, X. Dong, F. Xue, F. Y. Wang, Q. H. Wang and Z. H. Meng, Molecularly Imprinted Polymers for the Sensing of Explosives and Chemical Warfare Agents, Curr. Org. Chem., 2015, 19, 62–71 CrossRef CAS.
- A. McCluskey, C. I. Holdsworth and M. C. Bowyer, Molecularly imprinted polymers (MIPs): sensing, an explosive new opportunity?, Org. Biomol. Chem., 2007, 5, 3233–3244 RSC.
- M. Zarejousheghani, P. Rahimi, H. Borsdorf, S. Zimmermann and Y. Joseph, Molecularly Imprinted Polymer-Based Sensors for Priority Pollutants, Sensors, 2021, 21, 23 Search PubMed.
- D. L. Xiao, Y. Jiang and Y. P. Bi, Molecularly imprinted polymers for the detection of illegal drugs and additives: a review, Microchim. Acta, 2018, 185, 20 CrossRef PubMed.
- Y. T. Wang, Z. Q. Zhang, V. Jain, J. J. Yi, S. Mueller, J. Sokolov, Z. X. Liu, K. Levon, B. Rigas and M. H. Rafailovich, Potentiometric sensors based on surface molecular imprinting: Detection of cancer biomarkers and viruses, Sens. Actuators, B, 2010, 146, 381–387 CrossRef CAS.
- S. E. Diltemiz, R. Kecili, A. Ersoz and R. Say, Molecular Imprinting Technology in Quartz Crystal Microbalance (QCM) Sensors, Sensors, 2017, 17, 19 Search PubMed.
- M. C. Niu, P. H. Chuong and H. He, Core–shell nanoparticles coated with molecularly imprinted polymers: a review, Microchim. Acta, 2016, 183, 2677–2695 CrossRef CAS.
- J. Wackerlig and P. A. Lieberzeit, Molecularly imprinted polymer nanoparticles in chemical sensing – Synthesis, characterisation and application, Sens. Actuators, B, 2015, 207, 144–157 CrossRef CAS.
- J. Wackerlig and R. Schirhagl, Applications of Molecularly Imprinted Polymer Nanoparticles and Their Advances toward Industrial Use: A Review, Anal. Chem., 2016, 88, 250–261 CrossRef CAS PubMed.
- O. Hayden and F. L. Dickert, Selective microorganism detection with cell surface imprinted polymers, Adv. Mater., 2001, 13, 1480 CrossRef CAS.
- O. Ramstrom and R. J. Ansell, Molecular imprinting technology: Challenges and prospects for the future, Chirality, 1998, 10, 195–209 CrossRef CAS.
- E. Verheyen, J. P. Schillemans, M. van Wijk, M. A. Demeniex, W. E. Hennink and C. F. van Nostrum, Challenges for the effective molecular imprinting of proteins, Biomaterials, 2011, 32, 3008–3020 CrossRef CAS PubMed.
- C. Unger and P. A. Lieberzeit, Molecularly imprinted thin film surfaces in sensing: Chances and challenges, React. Funct. Polym., 2021, 161, 10 CrossRef.
- Y. He and Z. Lin, Recent advances in protein-imprinted polymers: synthesis, applications and challenges, J. Mater. Chem. B, 2022, 10, 6571–6589 RSC.
- D. Y. Choi, J. C. Yang and J. Park, Optimization and characterization of electrochemical protein Imprinting on hemispherical porous gold patterns for the detection of trypsin, Sens. Actuators, B, 2022, 350, 130855 CrossRef CAS.
- T. G. Halvorsen and L. Reubsaet, The utility of molecularly imprinted polymers for mass spectrometric protein and proteomics analysis, Proteomics, 2022, 22, 2100395 CrossRef CAS PubMed.
- W. Zhang, Y. Zhang, R. Wang, P. Zhang, Y. Zhang, E. Randell, M. Zhang and Q. Jia, A review: Development and application of surface molecularly imprinted polymers toward amino acids, peptides, and proteins, Anal. Chim. Acta, 2022, 1234, 340319 CrossRef CAS PubMed.
- Y. Guo, Y. Zheng, Y. Liu, X. Feng, Q. Dong, J. Li, J. Wang and C. Zhao, A concise detection strategy of Staphylococcus aureus using N-Succinyl-Chitosan-dopped bacteria-imprinted composite film and AIE fluorescence sensor, J. Hazard. Mater., 2022, 423, 126934 CrossRef CAS PubMed.
- L. Wang, Y. Nie, P. Wang, Y. Li, Q. Ma and D. Yu, A novel bacterial imprinted polymers- electrochemiluminescent sensor for Lactobacillus salivarius detection, Sens. Actuators, B, 2022, 358, 131467 CrossRef CAS.
- M. Harijan, V. Shukla, A. K. Singh, R. Raghuwanshi, G. Nath and M. Singh, Design of molecularly imprinted sensor for detection of typhoid using immunoinformatics and molecular imprinting, Biosens. Bioelectron.: X, 2022, 10, 100090 CAS.
- B. Bräuer, F. Thier, M. Bittermann, D. Baurecht and P. A. Lieberzeit, Raman Studies on Surface-Imprinted Polymers to Distinguish the Polymer Surface, Imprints, and Different Bacteria, ACS Appl. Bio Mater., 2022, 5, 160–171 CrossRef PubMed.
- S. Tokonami, Y. Nakadoi, H. Nakata, S. Takami, T. Kadoma, H. Shiigi and T. Nagaoka, Recognition of gram-negative and gram-positive bacteria with a functionalized conducting polymer film, Res. Chem. Intermed., 2014, 40, 2327–2335 CrossRef CAS.
- H. Bao, B. Yang, X. Zhang, L. Lei and Z. Li, Bacteria-templated fabrication of a charge heterogeneous polymeric interface for highly specific bacterial recognition, Chem. Commun., 2017, 53, 2319–2322 RSC.
- A. K. Yadav, D. Verma, N. Dalal, A. Kumar and P. R. Solanki, Molecularly imprinted polymer-based nanodiagnostics for clinically pertinent bacteria and virus detection for future pandemics, Biosens. Bioelectron.: X, 2022, 12, 100257 CAS.
- S. A. Khalid, R. Y. A. Hassan, R. M. El Nashar and I. M. El-Sherbiny, Voltammetric determination of Salmonella typhimurium in minced beef meat using a chip-based imprinted sensor, RSC Adv., 2022, 12, 3445–3453 RSC.
- R. Wang, L. Wang, J. Yan, D. Luan, s. Tao, J. Wu and X. Bian, Rapid, sensitive and label-free detection of pathogenic bacteria using a bacteria-imprinted conducting polymer film-based electrochemical sensor, Talanta, 2021, 226, 122135 CrossRef CAS PubMed.
- J. Pan, W. Chen, Y. Ma and G. Pan, Molecularly imprinted polymers as receptor mimics for selective cell recognition, Chem. Soc. Rev., 2018, 47, 5574–5587 RSC.
- N. Idil, M. Bakhshpour, I. Perçin and B. Mattiasson, Whole Cell Recognition of Staphylococcus aureus Using Biomimetic SPR Sensors, Biosensors, 2021, 11, 140 CrossRef CAS PubMed.
- E. Yilmaz, D. Majidi, E. Ozgur and A. Denizli, Whole cell imprinting based Escherichia coli sensors: A study for SPR and QCM, Sens. Actuators, B, 2015, 209, 714–721 CrossRef CAS.
- S. Piletsky, F. Canfarotta, A. Poma, A. M. Bossi and S. Piletsky, Molecularly Imprinted Polymers for Cell Recognition, Trends Biotechnol., 2020, 38, 368–387 CrossRef CAS PubMed.
- L. Dery, D. Zelikovich and D. Mandler, Electrochemistry of molecular imprinting of large entities, Curr. Opin. Electrochem., 2022, 34, 100967 CrossRef CAS.
- H. Gong, S. Hajizadeh, W. Liu and L. Ye, Imprinted Polymer Beads Loaded with Silver Nanoparticles for Antibacterial Applications, ACS Appl. Bio Mater., 2021, 4, 2829–2838 CrossRef CAS PubMed.
- A. Adan, G. Alizada, Y. Kiraz, Y. Baran and A. Nalbant, Flow cytometry: basic principles and applications, Crit. Rev. Biotechnol., 2017, 37, 163–176 CrossRef CAS PubMed.
- W. F. Wright, P. J. Simner, K. C. Carroll and P. G. Auwaerter, Progress Report: Next-Generation Sequencing, Multiplex Polymerase Chain Reaction, and Broad-Range Molecular Assays as Diagnostic Tools for Fever of Unknown Origin Investigations in Adults, Clin. Infect. Dis., 2022, 74, 924–932 CrossRef CAS PubMed.
- S. D. Gan and K. R. Patel, Enzyme Immunoassay and Enzyme-Linked Immunosorbent Assay, J. Invest. Dermatol., 2013, 133, E10–E12 CrossRef PubMed.
- V. Gubala, L. J. Johnston, Z. Liu, H. Krug, C. J. Moore, C. K. Ober, M. Schwenk and M. Vert, Engineered nanomaterials and human health: Part 1. Preparation, functionalization and characterization (IUPAC Technical Report), Pure Appl. Chem., 2018, 90, 1283–1324 CrossRef CAS.
- J. Zhang, Y. Hao, X. Tian, Y. Liang, X. He, R. Gao, L. Chen and Y. Zhang, Multi-stimuli responsive molecularly imprinted nanoparticles with tailorable affinity for modulated specific recognition of human serum albumin, J. Mater. Chem. B, 2022, 10, 6634–6643 RSC.
- Y. Toyoshima, A. Kawamura, Y. Takashima and T. Miyata, Design of molecularly imprinted hydrogels with thermoresponsive drug binding sites, J. Mater. Chem. B, 2022, 10, 6644–6654 RSC.
- A. Oshita, H. Sunayama and T. Takeuchi, A molecularly imprinted nanocavity with transformable domains that fluorescently indicate the presence of antibiotics in meat extract samples, J. Mater. Chem. B, 2022, 10, 6682–6687 RSC.
- X. Xue, M. Zhang, H. Gong and L. Ye, Recyclable nanoparticles based on a boronic acid–diol complex for the real-time monitoring of imprinting, molecular recognition and copper ion detection, J. Mater. Chem. B, 2022, 10, 6698–6706 RSC.
- C. Hong, J. Wang, Y. Wang, Z. Huang, H. Yang, D. Yang, R. Cai and W. Tan, Fluorescence detection of milk allergen β-lactoglobulin based on aptamers and WS2 nanosheets, J. Mater. Chem. B, 2022, 10, 6752–6757 RSC.
- A. Cumbo, B. Lorber, P. F.-X. Corvini, W. Meier and P. Shahgaldian, A synthetic nanomaterial for virus recognition produced by surface imprinting, Nat. Commun., 2013, 4, 1–7 Search PubMed.
- P. Rajapaksha, A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino and J. Chapman, A review of methods for the detection of pathogenic microorganisms, Analyst, 2019, 144, 396–411 RSC.
- L. Zeng, L. Wang and J. Hu, Current and emerging technologies for rapid detection of pathogens, Biosensing Technologies for the Detection of Pathogens-A Prospective Way for Rapid Analysis, 2018, vol. 73178, pp. 6–19 Search PubMed.
- W. S. Yoo, H. S. Han, J. G. Kim, K. Kang, H.-S. Jeon, J.-Y. Moon and H. Park, Development of a tablet PC-based portable device for colorimetric determination of assays including COVID-19 and other pathogenic microorganisms, RSC Adv., 2020, 10, 32946–32952 RSC.
- A. Soto, D. M. Quiñones-Laveriano, F. Valdivia, E. Juscamayta-López, J. Azañero-Haro, L. Chambi, H. Horna, G. Patiño, E. Guzman and J. A. De la Cruz-Vargas, Detection of viral and bacterial respiratory pathogens identified by molecular methods in COVID-19 hospitalized patients and its impact on mortality and unfavorable outcomes, Infect. Drug Resist., 2021, 14, 2795 CrossRef PubMed.
- A. K. Farha, Q.-Q. Yang, G. Kim, H.-B. Li, F. Zhu, H.-Y. Liu, R.-Y. Gan and H. Corke, Tannins as an alternative to antibiotics, Food Biosci., 2020, 38, 100751 CrossRef CAS.
- X. Zhao, Y. Zhang, J. Shi, Y. Liu, Y. Lu and Z. Lian, Biosynthesis of antibacterial compound against multidrug resistant foodborne pathogens by Phomopsis sp. XP-8, Food Control, 2019, 95, 223–231 CrossRef CAS.
- C. B. Chigor, I.-A. I. Ibangha, N. O. Nweze, V. C. Onuora, C. A. Ozochi, Y. Titilawo, M. C. Enebe, T. N. Chernikova, P. N. Golyshin and V. N. Chigor, Prevalence of integrons in multidrug-resistant Escherichia coli isolates from waters and vegetables in Nsukka and Enugu, Southeast Nigeria, Environ. Sci. Pollut. Res., 2022, 1–8 Search PubMed.
- S. T. Odonkor and K. K. Addo, Prevalence of multidrug-resistant Escherichia coli isolated from drinking water sources, Int. J. Microbiol., 2018, 2018 Search PubMed.
- Q. Ali, H. Zheng, M. J. Rao, M. Ali, A. Hussain, M. H. Saleem, Y. Nehela, M. A. Sohail, A. M. Ahmed and K. A. Kubar, Advances, limitations, and prospects of biosensing technology for detecting phytopathogenic bacteria, Chemosphere, 2022, 133773 CrossRef CAS PubMed.
- S. A. Zaidi, Bacterial imprinting methods and their applications: an overview, Crit. Rev. Anal. Chem., 2021, 51, 609–618 CAS.
- L. Wang, Y. Nie, P. Wang, Y. Li, Q. Ma and D. Yu, A novel bacterial imprinted polymers-electrochemiluminescent sensor for Lactobacillus salivarius detection, Sens. Actuators, B, 2022, 358, 131467 CrossRef CAS.
- M. S. Amorim, M. G. F. Sales and M. F. Frasco, Recent advances in virus imprinted polymers, Biosens. Bioelectron.: X, 2022, 100131 CAS.
- G. J. Soufi, S. Iravani and R. S. Varma, Molecularly imprinted polymers for the detection of viruses: Challenges and opportunities, Analyst, 2021, 146, 3087–3100 RSC.
- N. M. Bergmann and N. A. Peppas, Molecularly imprinted polymers with specific recognition for macromolecules and proteins, Prog. Polym. Sci., 2008, 33, 271–288 CrossRef CAS.
- F. Eisenhaber, B. Persson and P. Argos, Protein structure prediction: recognition of primary, secondary, and tertiary structural features from amino acid sequence, Crit. Rev. Biochem. Mol. Biol., 1995, 30, 1–94 CrossRef CAS PubMed.
- Y. Lv, T. Tan and F. Svec, Molecular imprinting of proteins in polymers attached to the surface of nanomaterials for selective recognition of biomacromolecules, Biotechnol. Adv., 2013, 31, 1172–1186 CrossRef CAS PubMed.
- J. D. Hoff, L.-J. Cheng, E. Meyhöfer, L. J. Guo and A. J. Hunt, Nanoscale protein patterning by imprint lithography, Nano Lett., 2004, 4, 853–857 CrossRef CAS.
- L. Karadurmus, M. E. Corman, L. Uzun and S. A. Ozkan, Enantioselective recognition of esomeprazole with a molecularly imprinted sol–gel-based electrochemical sensor, Microchim. Acta, 2022, 189, 1–9 CrossRef PubMed.
- F. Yin, Y. Mo, X. Liu, Y. Pang, X. Wu, L. Hao, J. Yu and F. Xu, Surface-imprinted polymer microspheres for rapid and selective adsorption of As (V) ions from the aqueous phase, Mater. Chem. Phys., 2022, 281, 125687 CrossRef CAS.
- S. Akhtarian, A. Doostmohammadi, K. Youssef, G. Kraft, S. Kaur Brar and P. Rezai, Metal Microwires Functionalized with Cell-Imprinted Polymer for Capturing Bacteria in Water, ACS Appl. Polym. Mater., 2023, 55, 3235–3246 CrossRef.
- J. Zhang, Y. Wang and X. Lu, Molecular imprinting technology for sensing foodborne pathogenic bacteria, Anal. Bioanal. Chem., 2021, 413, 4581–4598 CrossRef CAS PubMed.
- F. Cui, Z. Zhou and H. S. Zhou, Molecularly Imprinted Polymers and Surface Imprinted Polymers Based Electrochemical Biosensor for Infectious Diseases, Sensors, 2020, 20, 996 CrossRef CAS PubMed.
- R. Tian, Y. Li, J. Xu, C. Hou, Q. Luo and J. Liu, Recent development in the design of artificial enzymes through molecular imprinting technology, J. Mater. Chem. B, 2022, 10, 6590–6606 RSC.
- N. Idil and B. Mattiasson, Imprinting of Microorganisms for Biosensor Applications, Sensors, 2017, 17, 708 CrossRef PubMed.
- M. Jia, Z. Zhang, J. Li, X. Ma, L. Chen and X. Yang, Molecular imprinting technology for microorganism analysis, TrAC, Trends Anal. Chem., 2018, 106, 190–201 CrossRef CAS.
- C. Dong, H. Shi, Y. Han, Y. Yang, R. Wang and J. Men, Molecularly imprinted polymers by the surface imprinting technique, Eur. Polym. J., 2021, 145, 110231 CrossRef CAS.
- M. Werner, M. S. Glück, B. Bräuer, A. Bismarck and P. A. Lieberzeit, Investigations on sub-structures within cavities of surface imprinted polymers using AFM and PF-QNM, Soft Matter, 2022, 18, 2245–2251 RSC.
- B. Bräuer, F. Thier, M. Bittermann, D. Baurecht and P. A. Lieberzeit, Raman Studies on Surface-Imprinted Polymers to Distinguish the Polymer Surface, Imprints, and Different Bacteria, ACS Appl. Bio Mater., 2021, 5, 160–171 CrossRef PubMed.
- V. Razavilar, H. Ahari, B. Akbari Adergani and A. Anvar, A central composite face-centered design for optimizing the detection of Salmonella typhi with a fluorescence nanobiosensor using the microcontact method, Int. J. Environ. Sci. Technol., 2019, 16, 4637–4646 CrossRef CAS.
- S. M. Mugo, W. Lu and D. Dhanjai, A pathogen imprinted hybrid polymer capacitive sensor for selective Escherichia coli detection, Med. Devices Sens., 2020, 3, e10071 CrossRef.
- G. Ertürk and R. Lood, Bacteriophages as biorecognition elements in capacitive biosensors: Phage and host bacteria detection, Sens. Actuators, B, 2018, 258, 535–543 CrossRef.
- B. van Grinsven, K. Eersels, O. Akkermans, S. Ellermann, A. Kordek, M. Peeters, O. Deschaume, C. Bartic, H. Diliën and E. Steen Redeker, Label-free detection of Escherichia coli based on thermal transport through surface imprinted polymers, ACS Sens., 2016, 1, 1140–1147 CrossRef CAS.
- P. Cornelis, S. Givanoudi, D. Yongabi, H. Iken, S. Duwé, O. Deschaume, J. Robbens, P. Dedecker, C. Bartic and M. Wübbenhorst, Sensitive and specific detection of E. coli using biomimetic receptors in combination with a modified heat-transfer method, Biosens. Bioelectron., 2019, 136, 97–105 CrossRef CAS PubMed.
- A. Karthik, K. Margulis, K. Ren, R. N. Zare and L. W. Leung, Rapid and selective detection of viruses using virus-imprinted polymer films, Nanoscale, 2015, 7, 18998–19003 RSC.
- A. Seifner, P. Lieberzeit, C. Jungbauer and F. L. Dickert, Synthetic receptors for selectively detecting erythrocyte ABO subgroups, Anal. Chim. Acta, 2009, 651, 215–219 CrossRef CAS PubMed.
- R. Samardzic, H. F. Sussitz, N. Jongkon and P. A. Lieberzeit, Quartz crystal microbalance in-line sensing of escherichia coli in a bioreactor using molecularly imprinted polymers, Sens. Lett., 2014, 12, 1152–1155 CrossRef.
- J. Starosvetsky, T. Cohen, U. Cheruti, D. Dragoljub and R. Armon, Effects of physical parameters on bacterial cell adsorption onto pre-imprinted sol-gel films, J. Biomater. Nanobiotechnol., 2012, 3, 499 CrossRef.
- R. Schirhagl, E. W. Hall, I. Fuereder and R. N. Zare, Separation of bacteria with imprinted polymeric films, Analyst, 2012, 137, 1495–1499 RSC.
- Y. Wang, Z. Zhang, V. Jain, J. Yi, S. Mueller, J. Sokolov, Z. Liu, K. Levon, B. Rigas and M. H. Rafailovich, Potentiometric sensors based on surface molecular imprinting: Detection of cancer biomarkers and viruses, Sens. Actuators, B, 2010, 146, 381–387 CrossRef CAS.
- T. Zhou, K. Zhang, T. Kamra, L. Bülow and L. Ye, Preparation of protein imprinted polymer beads by Pickering emulsion polymerization, J. Mater. Chem. B, 2015, 3, 1254–1260 RSC.
- X. Zhao, Y. Cui, J. Wang and J. Wang, Preparation of fluorescent molecularly imprinted polymers via pickering emulsion interfaces and the application for visual sensing analysis of Listeria Monocytogenes, Polymers, 2019, 11, 984 CrossRef CAS PubMed.
- X. Shen, J. Svensson Bonde, T. Kamra, L. Bülow, J. C. Leo, D. Linke and L. Ye, Bacterial imprinting at Pickering emulsion interfaces, Angew. Chem., Int. Ed., 2014, 53, 10687–10690 CrossRef CAS PubMed.
- S. A. Khalid, R. Y. Hassan, R. M. El Nashar and I. M. El-Sherbiny, Voltammetric determination of Salmonella typhimurium in minced beef meat using a chip-based imprinted sensor, RSC Adv., 2022, 12, 3445–3453 RSC.
- R. Wang, L. Wang, J. Yan, D. Luan, J. Wu and X. Bian, Rapid, sensitive and label-free detection of pathogenic bacteria using a bacteria-imprinted conducting polymer film-based electrochemical sensor, Talanta, 2021, 226, 122135 CrossRef CAS PubMed.
- N. Yasmeen, M. Etienne, P. S. Sharma, S. El-Kirat-Chatel, M. B. Helú and W. Kutner, Molecularly imprinted polymer as a synthetic receptor mimic for capacitive impedimetric selective recognition of Escherichia coli K-12, Anal. Chim. Acta, 2021, 1188, 339177 CrossRef CAS PubMed.
- S. Chen, X. Chen, L. Zhang, J. Gao and Q. Ma, Electrochemiluminescence detection of Escherichia coli O157: H7 based on a novel polydopamine surface imprinted polymer biosensor, ACS Appl. Mater. Interfaces, 2017, 9, 5430–5436 CrossRef CAS PubMed.
- A. A. Lahcen, F. Arduini, F. Lista and A. Amine, Label-free electrochemical sensor based on spore-imprinted polymer for Bacillus cereus spore detection, Sens. Actuators, B, 2018, 276, 114–120 CrossRef.
- R. D. Crapnell, A. Hudson, C. W. Foster, K. Eersels, B. v. Grinsven, T. J. Cleij, C. E. Banks and M. Peeters, Recent advances in electrosynthesized molecularly imprinted polymer sensing platforms for bioanalyte detection, Sensors, 2019, 19, 1204 CrossRef CAS PubMed.
- G. Pilvenyte, V. Ratautaite, R. Boguzaite, U. Samukaite-Bubniene, D. Plausinaitis, A. Ramanaviciene, M. Bechelany and A. Ramanavicius, Molecularly imprinted polymers for the recognition of biomarkers of certain neurodegenerative diseases, J. Pharm. Biomed. Anal., 2023, 228, 115343 CrossRef CAS PubMed.
- Z. Zhang, Y. Guan, M. Li, A. Zhao, J. Ren and X. Qu, Highly stable and reusable imprinted artificial antibody used for in situ detection and disinfection of pathogens, Chem. Sci., 2015, 6, 2822–2826 RSC.
- J. Borovička, S. D. Stoyanov and V. N. Paunov, Shape recognition of microbial cells by colloidal cell imprints, Nanoscale, 2013, 5, 8560–8568 RSC.
- J. Borovička, W. J. Metheringham, L. A. Madden, C. D. Walton, S. D. Stoyanov and V. N. Paunov, Photothermal colloid antibodies for shape-selective recognition and killing of microorganisms, J. Am. Chem. Soc., 2013, 135, 5282–5285 CrossRef PubMed.
- A. Findeisen, J. Wackerlig, R. Samardzic, J. Pitkänen, O. Anttalainen, F. L. Dickert and P. A. Lieberzeit, Artificial receptor layers for detecting chemical and biological agent mimics, Sens. Actuators, B, 2012, 170, 196–200 CrossRef CAS.
- C. J. Tan and Y. W. Tong, The effect of protein structural conformation on nanoparticle molecular imprinting of ribonuclease a using miniemulsion polymerization, Langmuir, 2007, 23, 2722–2730 CrossRef CAS PubMed.
- B. Pluhar, U. Ziener and B. Mizaikoff, Binding performance of pepsin surface-imprinted polymer particles in protein mixtures, J. Mater. Chem. B, 2015, 3, 6248–6254 RSC.
- S. M. DePorter, I. Lui and B. R. McNaughton, Programmed cell adhesion and growth on cell-imprinted polyacrylamide hydrogels, Soft Matter, 2012, 8, 10403–10408 RSC.
- P. Qi, Y. Wan and D. Zhang, Impedimetric biosensor based on cell-mediated bioimprinted films for bacterial detection, Biosens. Bioelectron., 2013, 39, 282–288 CrossRef CAS PubMed.
- Z. Altintas, J. Pocock, K.-A. Thompson and I. E. Tothill, Comparative investigations for adenovirus recognition and quantification: Plastic or natural antibodies?, Biosens. Bioelectron., 2015, 74, 996–1004 CrossRef CAS PubMed.
- X. Xue, J. Pan, H. Xie, J. Wang and S. Zhang, Specific recognition of staphylococcus aureus by staphylococcus aureus protein A-imprinted polymers, React. Funct. Polym., 2009, 69, 159–164 CrossRef CAS.
- M. A. R. Khan, F. T. C. Moreira, J. Riu and M. G. F. Sales, Plastic antibody for the electrochemical detection of bacterial surface proteins, Sens. Actuators, B, 2016, 233, 697–704 CrossRef CAS.
- R. Schirhagl, K. Ren and R. N. Zare, Surface-imprinted polymers in microfluidic devices, Sci. China: Chem., 2012, 55, 469–483 CrossRef CAS.
- S. Sykora, A. Cumbo, G. Belliot, P. Pothier, C. Arnal, Y. Dudal, P. F. X. Corvini and P. Shahgaldian, Virus-like particles as virus substitutes to design artificial virus-recognition nanomaterials, Chem. Commun., 2015, 51, 2256–2258 RSC.
- R. Hu, J. Luan, E. D. Kharasch, S. Singamaneni and J. J. Morrissey, Aromatic Functionality of Target Proteins Influences Monomer Selection for Creating Artificial Antibodies on Plasmonic Biosensors, ACS Appl. Mater. Interfaces, 2017, 9, 145–151 CrossRef CAS PubMed.
- S. Kraus-Ophir, J. Witt, G. Wittstock and D. Mandler, Nanoparticle-Imprinted Polymers for Size-Selective Recognition of Nanoparticles, Angew. Chem., Int. Ed., 2014, 53, 294–298 CrossRef CAS PubMed.
- J. Witt, D. Mandler and G. Wittstock, Nanoparticle-Imprinted Matrices as Sensing Layers for Size-Selective Recognition of Silver Nanoparticles, ChemElectroChem, 2016, 3, 2116–2124 CrossRef CAS.
- M. K. Corbierre, N. S. Cameron, M. Sutton, S. G. J. Mochrie, L. B. Lurio, A. Rühm and R. B. Lennox, Polymer-Stabilized Gold Nanoparticles and Their Incorporation into Polymer Matrices, J. Am. Chem. Soc., 2001, 123, 10411–10412 CrossRef CAS PubMed.
- A. B. R. Mayer, Formation of noble metal nanoparticles within a polymeric matrix: nanoparticle features and overall morphologies, Mater. Sci. Eng., C, 1998, 6, 155–166 CrossRef.
- S. Bharathi and O. Lev, Direct synthesis of gold nanodispersions in sol–gel derived silicate sols, gels and films, Chem. Commun., 1997, 2303–2304, 10.1039/A705609E.
- S. C. Biradar, D. B. Shinde, V. K. Pillai and M. G. Kulkarni, Polydentate disulfides for enhanced stability of AuNPs and facile nanocavity formation, J. Mater. Chem., 2012, 22, 10000–10008 RSC.
- S. Koenig and V. Chechik, Au nanoparticle-imprinted polymers, Chem. Commun., 2005, 4110–4112, 10.1039/B503243A.
- S. Gam-Derrouich, C. Bourdillon, W. Daney De Marcillac, L. Coolen, A. Maître, C. Mangeney and C. Schwob, Quantum dot-imprinted polymers with size and shell-selective recognition properties, Chem. Commun., 2015, 51, 14933–14936 RSC.
- X. Chen, M. Hirtz, H. Fuchs and L. Chi, Fabrication of Gradient Mesostructures by Langmuir–Blodgett Rotating Transfer, Langmuir, 2007, 23, 2280–2283 CrossRef CAS PubMed.
- S. Watanabe, H. Shibata, F. Sakamoto, R. Azumi, H. Sakai, M. Abe and M. Matsumoto, Directed self-assembly of gold nanoparticles and gold thin films on micro- and nanopatterned templates fabricated from mixed phase-separated Langmuir-Blodgett films, J. Mater. Chem., 2009, 19, 6796–6803 RSC.
- S. Acharya, B. Das, U. Thupakula, K. Ariga, D. D. Sarma, J. Israelachvili and Y. Golan, A Bottom-Up Approach toward Fabrication of Ultrathin PbS Sheets, Nano Lett., 2013, 13, 409–415 CrossRef CAS PubMed.
- J. Benito, S. Sorribas, I. Lucas, J. Coronas and I. Gascon, Langmuir–Blodgett Films of the Metal–Organic Framework MIL-101(Cr): Preparation, Characterization, and CO2 Adsorption Study Using a QCM-Based Setup, ACS Appl. Mater. Interfaces, 2016, 8, 16486–16492 CrossRef CAS PubMed.
- D. I. Babenko, A. A. Ezhov, D. S. Turygin, V. K. Ivanov, V. V. Arslanov and M. A. Kalinina, 2D “Soap”-Assembly of Nanoparticles via Colloid-Induced Condensation of Mixed Langmuir Monolayers of Fatty Surfactants, Langmuir, 2012, 28, 125–133 CrossRef CAS PubMed.
- J. Huang, A. R. Tao, S. Connor, R. He and P. Yang, A General Method for Assembling Single Colloidal Particle Lines, Nano Lett., 2006, 6, 524–529 CrossRef CAS PubMed.
- P. Pienpinijtham, X. X. Han, S. Ekgasit and Y. Ozaki, An ionic surfactant-mediated Langmuir–Blodgett method to construct gold nanoparticle films for surface-enhanced Raman scattering, Phys. Chem. Chem. Phys., 2012, 14, 10132–10139 RSC.
- S. Sun, P. H. Ho-Si and D. J. Harrison, Preparation of active Langmuir-Blodgett films of glucose oxidase, Langmuir, 1991, 7, 727–737 CrossRef CAS.
- N. Bruchiel-Spanier and D. Mandler, Nanoparticle-Imprinted Polymers: Shell-Selective Recognition of Au Nanoparticles by Imprinting Using the Langmuir–Blodgett Method, ChemElectroChem, 2015, 2, 795–802 CrossRef CAS.
- D. Zelikovich, S. Dery, N. Bruchiel-Spanier, N. Tal, P. Savchenko, E. Gross and D. Mandler, Shell–Matrix Interaction in Nanoparticle-Imprinted Matrices: Implications for Selective Nanoparticle Detection and Separation, ACS Appl. Nano Mater., 2021, 4, 10819–10827 CrossRef CAS.
- N. Bruchiel-Spanier, L. Dery, N. Tal, S. Dery, E. Gross and D. Mandler, Effect of matrix-nanoparticle interactions on recognition of aryldiazonium nanoparticle-imprinted matrices, Nano Res., 2019, 12, 265–271 CrossRef CAS.
- N. Bruchiel-Spanier, G. Giordano, A. Vakahi, M. Guglielmi and D. Mandler, Electrochemically Deposited Sol–Gel Based Nanoparticle-Imprinted Matrices for the Size-Selective Detection of Gold Nanoparticles, ACS Appl. Nano Mater., 2018, 1, 5612–5619 CrossRef CAS.
- M. Hitrik, Y. Pisman, G. Wittstock and D. Mandler, Speciation of nanoscale objects by nanoparticle imprinted matrices, Nanoscale, 2016, 8, 13934–13943 RSC.
- D. Xia and S. R. J. Brueck, A Facile Approach to Directed Assembly of Patterns of Nanoparticles Using Interference Lithography and Spin Coating, Nano Lett., 2004, 4, 1295–1299 CrossRef CAS.
- D. Zelikovich, P. Savchenko and D. Mandler, High Recognition of Mercaptobenzoic Acid Isomer Stabilized Gold Nanoparticles through Matrix Imprinting, ACS Appl. Mater. Interfaces, 2023, 15, 32687–32696 CrossRef CAS PubMed.
- R. Shacham, D. Avnir and D. Mandler, Electrodeposition of Methylated Sol-Gel Films on Conducting Surfaces, Adv. Mater., 1999, 11, 384–388 CrossRef CAS.
- M. Sheffer, A. Groysman and D. Mandler, Electrodeposition of sol–gel films on Al for corrosion protection, Corros. Sci., 2003, 45, 2893–2904 CrossRef CAS.
- B. Veeraraghavan, B. Haran, D. Slavkov, S. Prabhu, B. Popov and B. Heimann, Development of a Novel Electrochemical Method to Deposit High Corrosion Resistant Silicate Layers on Metal Substrates, Electrochem. Solid-State Lett., 2003, 6, B4 CrossRef CAS.
- R. Shacham, D. Avnir and D. Mandler, Electrodeposition of Methylated Sol-Gel Films on Conducting Surfaces, Adv. Mater., 1999, 11, 384–388 CrossRef CAS.
- L. Dery, B. Sava and D. Mandler, Electrochemical Detection of Silica Nanoparticles by Nanoparticle Imprinted Matrices, ChemElectroChem, 2023, 10, e202300039 CrossRef CAS.
- L. Dery, N. Shauloff, Y. Turkulets, I. Shalish, R. Jelinek and D. Mandler, Size-Selective Detection of Nanoparticles in Solution and Air by Imprinting, ACS Sens., 2022, 7, 296–303 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |