Open Access Article
Open Access ArticleCongo red protects bacteriophages against UV irradiation and allows for the simultaneous use of phages and UV for membrane sterilization†
Mateusz
Wdowiak
 ,
Patryk A.
Mierzejewski
,
Rafał
Zbonikowski
,
Patryk A.
Mierzejewski
,
Rafał
Zbonikowski
 ,
Bartłomiej
Bończak
and
Jan
Paczesny
,
Bartłomiej
Bończak
and
Jan
Paczesny
 *
*
Institute of Physical Chemistry PAS, Kasprzaka 44/52, 01-224 Warsaw, Poland. E-mail: jpaczesny@ichf.edu.pl; Tel: +48 22 343 2071
First published on 16th January 2023
Abstract
The membrane bioreactor market has been expected to grow by 4.9 billion USD within five years – from 2021 to 2026. Biofouling caused by the deposition of microorganisms and their extracellular substances decreases the operational flux of the membrane bioreactor, increasing the energy consumption costs of bioreactor maintenance. Standard protocols are costly and time-consuming, and generate harmful waste. Here, a method for the simultaneous utilization of UV light and bacteriophages for efficient sterilization of bacteria-contaminated membranes is demonstrated. Congo red, a commonly used dye, provides selective protection of phages, but not bacteria, against UV radiation. This allows for overcoming the significant drawback of such protocols – the phage inactivation by UV. The simultaneous utilization of phages, UV, and Congo red resulted in the complete removal of bacteria from membranes (>4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log) within just 30 minutes. This is also the first attempt to describe the UV-protective property mechanism of dyes.
log) within just 30 minutes. This is also the first attempt to describe the UV-protective property mechanism of dyes.
Water impactApplying bacteriophages and Congo red (‘cleaning mixture’) for UV membrane sterilization significantly shortens the procedure time and decreases the number of chemicals used for bioreactor cleaning. Congo red has low toxicity (similar to caffeine) and can be easily removed from water solutions using different approaches. Therefore, applying a ‘cleaning mixture’ may limit the release of chemicals into water environments. |
Introduction
The global market for membrane bioreactors is estimated to grow to 4.9 billion USD with a compound annual growth rate (CAGR) of 8.3% by 2026. From 2018 to 2023, its growth was even faster, from a value of 1.9 billion USD to 3.8 billion USD within five years, with a CAGR of 14.7%.1 Another report estimated this sector to be 3.0 billion USD in 2019.2 Biofouling is regarded as one of the biggest threats to the membrane bioreactor industry. Compared to standard maintenance procedures, the cost of cleaning fouled membranes increases by 50–100%. Also, operating with fouled membranes increases the cost of energy or even causes a malfunction of the equipment.3Biofouling is most commonly treated with chemicals.4 However, they are often corrosive, which affects the membrane integrity. Disposing of used chemicals and by-products harms health, safety, and the environment.5 Another commonly used sterilization agent is UV light. Its application is suitable yet limited by the geometry of the bioreactors and hindered access to cloudy media.6 Other means are either costly (e.g., antibiotics or antifungal drugs) or time-consuming (like temperature-sterilization7), or require specialized equipment (e.g., ozonation8), including corrosion-resistant properties (e.g., for chlorine sterilization9). Therefore, there is a need to develop new methods to deal with membrane fouling. The preferred method would be based on mild agents that do not generate toxic wastes. Scarascia et al. proposed using UV and bacteriophages for two-step membrane sterilization.10 However, such an approach has an intrinsic drawback: UV deactivates bacteria and phages alike. Therefore, UV and phages cannot be used simultaneously.
Bacteriophages (phages) are viruses that infect bacteria. Phages are safe for humans,11 cheap, and produced using rapid and inexpensive techniques.12 The vast majority of phages cause the host's death upon completion of the reproduction cycle. Thus phages are considered natural antibacterial agents. This fact makes phages the most promising ‘tools’ in the fight against the growing antibiotic resistance among bacterial strains.13,14 The global bacteriophage market (i.e., phage therapies, food industry, agriculture) is forecasted to change with a CAGR of 4.5% in 2021–2029.15
The use of bacteriophages is an exciting solution to eradicate fouling agents such as bacterial cells and is of great interest for industrial applications. Such research is, however, in a preliminary stage. The specificity of particular phages to only certain bacteria requires the combination of bacteriophage sterilization with less specific bactericidal factors, e.g., UV radiation. Several reports have proven the superior efficacy of the two-step sterilization using first UV radiation and then bacteriophages.16–18 Recently, Scarascia et al.,10 for the first time, showed the additive effect of the consecutive application of UV and phages for efficient bioreactor membrane sterilization. The method was found significantly different, yet comparable to the efficacy of chemical treatment (the alive/dead cell ratio for chemical treatment was about 0.25, while after UV-C + bacteriophage treatment, the ratio was about 0.35). The authors compared the efficacy of the simultaneous use of UV and bacteriophages for 3 h and 6 h and bacteriophage exposure after UV exposure. Significant differences in the efficacy of bacteria elimination were observed during independent UV-C + bacteriophage and simultaneous UV-C + bacteriophage exposure. Although the simultaneous use of UV-C and bacteriophages appeared to be significantly more efficient than their independent application, the additive effect was smaller than could be expected. This might be due to the fact that ultraviolet radiation inactivates bacteriophages. UV radiation primarily targets the viral genome, with pyrimidine bases being more reactive than purine at the wavelength of 254 nm.19 This fact was confirmed for various phages (icosahedral, e.g., T2, PhiX174, and filamentous, e.g., MS2, fr)20 and animal viruses such as herpes simplex virus21 or poliovirus.22 Although some coliphages, such as T7, can develop UV resistance, this process is prolonged and inefficient.23
The method described by Scarascia et al. takes 3 or 6 hours,10 which means that such a protocol is significantly faster than chemical-based sterilization (96 hours for chlorine,24 up to a few days for ozonation25) with comparable efficacy. However, knowing the detrimental effect of UV on the phages, protection against highly energetic radiation becomes even more urgent.26 The simultaneous application of UV radiation and bacteriophages, protected from the adverse effects of UV, would result in an even more efficient and possibly quicker sterilization protocol. We found that Congo red (CR) dye can be used for such an application as it selectively protects bacteriophages from UV irradiation. Moreover, we confirmed the uniqueness of this dye by comparing CR to five other judiciously chosen dyes.
Congo red (CR for short), a representative of azo dyes, has been known for over 100 years as a staining dye. It is frequently used in medicine for amyloids and aggregated protein staining.27–30 The toxicity of Congo red to rats is estimated to be LD50 = 15.2 g per kilogram of body mass.27,31 A lethal dose for humans is estimated to be about 143 mg per kg of body mass, i.e., similar to caffeine (LD50 of 150–200 mg kg−1 (ref. 32)).
The protective action of CR towards bacteriophages was unexpected, as a significant body of literature has shown the opposite effects against other viruses. Rawal and Vyas proved the virucidal properties of Congo red (and ascorbic acid) to the human immunodeficiency virus (HIV).33 In the paper by Estupinan and Hanson, significantly decreased infectivity of the Newcastle disease virus (NDV) was observed when the virus was injected into chicken embryos simultaneously with Congo red.34 The inactivation of virions by CR (2.4 μg ml−1) is known not only for NDV but also for influenza virus, fowl plague virus, and Sindbis virus.34,35 The effect of CR on bacteriophages has not been investigated until now.
Here, a potential application of CR to protect the majority of bacteriophages against UV irradiation is proposed. Only in the case of the enveloped Phi 6 phage did Congo red prove to deactivate virions. This knowledge was used to develop a procedure for dealing with membrane fouling, which utilizes the simultaneous action of UV and bacteriophages. Congo red was proved to protect phages, but not Gram-negative bacteria, against UV under the same experimental conditions.
Experimental
1. Bacteriophage protection against UV radiation
10 mg of the dye was dissolved in 1 ml of TM buffer, reaching a final concentration of CR = 1% (w/v). During the experiment, 6 species of non-enveloped bacteriophages – T4 (Tevenviridae), T7 (Studierviridae), T1 (Tunaviridae), MS2 (Leviviridae), M13 (Inoviridae), and LR1_PA01 (environmental; tailed phage isolated from the Baltic seawater; probably Pbunavirus) were examined. These phages represented the groups of viruses with different types of genome organization – double-stranded DNA (dsDNA; T1, T4, T7), single-stranded DNA (ssDNA; M13), and single-stranded RNA ((+)ssRNA; MS2). As a representative of enveloped bacteriophages, Pseudomonas phage Phi 6 (DSM 21518; obtained from German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) was used. This phage is a member of the Cystoviridae family, with the genome in the form of double-stranded RNA (dsRNA). As the host for the T1, T4, and T7 phages, the following strain was used: Escherichia coli BL21 strain; for the MS2 and M13 phages, the host was the E. coli C3000 strain; for LR1_PA01, we used the Pseudomonas aeruginosa PAO1 strain; for the Phi 6 phage, we used the Pseudomonas syringae DSM 21482 strain (obtained from German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). All the examined phages were suspended in the TM buffer solution (with or without the addition of CR) to reach the initial concentration of 105 PFU ml−1 (plaque-forming units per milliliter). Every prepared specimen was incubated at 4 °C for 24 h.After 24 hours of incubation, titration was performed by a droplet test on double-layer LB-agar plates. The top-agar layer contained host cells of E. coli BL21 (T1, T4, T7 phages), E. coli C3000 (MS2, M13 phages), and P. aeruginosa PAO1 (LR1_PA01), or P. syringae DSM 21482 (Phi 6) strains. After the incubation with CR, a droplet test was performed by placing at least eight droplets (5 μl each) of each phage suspension on the top-agar layer.
The remaining bulk of each sample was transferred to sterile glass Petri dishes (90 mm diameter). The use of plastic Petri dishes might cause the generation of oxygen radicals from plastic elements,36 cause the uncontrolled adsorption of phages,37 and thus affect the results of the experiments. Irradiation was performed by placing glass Petri dishes inside a UV chamber (CL-1000 Ultraviolet Crosslinker, UVP; 5 × 8 W, 254 nm UV) for 1 minute or 60 minutes (depending on the experiment) with plates positioned 15 cm from the radiation source.
The second titration took place after all the samples were exposed to UVC radiation (ca. 100–280 nm wavelength) for 1 minute. A total bacteriophage count was performed after either 4–5 hours (for T1 and T7) or overnight incubation (for MS2, M13, T4, and LR1_PA01 and Phi 6). All experiments were performed in triplicate.
2. Bacteria protection against UV
E. coli BL21 (obtained from the Institute of Biochemistry and Biophysics in Warsaw, Poland) and P. aeruginosa PAO1 (obtained from the collection of the Institute of Physical Chemistry PAS) were used as representatives of Gram-negative strains. Listeria monocytogenes EGDe (obtained from the University of Warsaw Biological and Chemical Research Centre in Warsaw, Poland) and Enterococcus durans PCM1857 strains (obtained from Polish Collection of Microorganisms) were used as Gram-positive bacteria representatives. The bacteria were cultured according to the standard protocols. The single colony from the culture on the LB agar (E. coli, P. aeruginosa, E. durans) or BHI agar (L. monocytogenes) plate was inoculated into 10 ml of LB or BHI medium and then incubated at 37 °C in an orbital shaker (220 rpm) for ca. 16 hours (overnight culture). Next, the overnight cultures were refreshed by adding 7.5 ml of fresh LB or BHI medium to 2.5 ml of overnight culture and incubated for one hour at 37 °C. Then, the refreshed cultures were both diluted with LB or BHI medium to reach an optical density of around OD600 = 0.8 for E. coli (this value corresponds to about 6 × 108 CFU ml−1 (colony-forming units per ml)) or OD600 = 0.9 for P. aeruginosa (which corresponds to about 2.5 × 108 CFU ml−1), OD600 = 1.06 for L. monocytogenes (this value corresponds to about 1 × 1010 CFU ml−1), or OD600 = 0.65 for E. durans (this corresponds to about 0.5 × 1010 CFU ml−1). After that, the bacteria were diluted in 0.9% NaCl solution or 0.9% NaCl with 1% Congo red (w/v) to reach the initial concentration of 104 CFU ml−1. Before 24 hours of incubation at room temperature with shaking (400 rpm), 100 μL of each solution was taken to be cultured on LB or BHI agar plates. After incubation, another 100 μL of bacteria solution was cultured on LB or BHI agar plates to investigate if Congo red affected bacterial growth. The remaining samples were transferred to sterile glass plates and exposed to UV-C radiation for a particular period (1 minute) inside the UV chamber (CL-1000 Ultraviolet Crosslinker, UVP; 5 × 8 W, 254 nm UV). After the exposure, 100 μl of the bacterial solution was cultured on LB or BHI agar plates. In each case, the plate count method was used to determine the viability of the bacteria.Finally, a mixture of the LR1_PA01 bacteriophage and E. coli BL21 bacteria was prepared. The initial concentration of the LR1_PA01 phage in the TM buffer was about 105 PFU ml−1, and the initial concentration of E. coli in 0.9% NaCl was about 105 CFU ml−1. The experimental set also contained 1% of Congo red. After the preparation, 100 μL of each mixture was cultured on LB-agar plates. A droplet test was performed simultaneously – eight droplets (5 μl) of the mixtures diluted 1, 10, and 100 times were deposited on double-layer LB-agar plates with P. aeruginosa PAO1 bacteria (the host bacteria for the LR1_PA01 phage) in the top-agar layer. The mixtures were incubated at room temperature with shaking (400 rpm) for 24 hours. After that, droplet and plating tests were performed, and then the bacteria and phages were exposed to UV radiation for 1 minute. After the exposure, the droplet and plating tests were performed again. All experiments were conducted in triplicate.
3. Yeast stabilization against UV
The Saccharomyces cerevisiae WT (wild type) strain (obtained from the Institute of Biochemistry and Biophysics in Warsaw, Poland) was used as an example of yeasts. The yeasts were cultured according to the standard protocol. The single colony from the culture on a YPD agar plate was inoculated into 10 ml of YPD medium and then incubated at 37 °C in an orbital shaker (220 rpm) for ca. 16 hours (overnight culture). Next, the overnight cultures were refreshed by adding 7.5 ml of fresh YPD medium to 2.5 ml of overnight culture and incubated for one hour at 37 °C. Then, the refreshed cultures were diluted with YPD medium to reach an optical density of OD600 = 0.75, corresponding to a concentration of about 1 × 107 cells per ml. Afterward, the yeasts were diluted in 0.9% NaCl solution or 0.9% NaCl with 1% Congo red (w/v) to reach the initial concentration of 103 cells per ml. Before 24 hours of incubation at room temperature with shaking (400 rpm), 100 μL of each solution was plated onto YPD agar plates. After incubation, another 100 μL of S. cerevisiae solution was taken to be cultured on YPD agar plates to investigate if CR affected yeast growth. The samples were transferred to sterile glass plates and exposed to UVC radiation for 1 minute inside the UV chamber (CL-1000 Ultraviolet Crosslinker, UVP; 5 × 8 W, 254 nm UV). Exposure conditions were the same as those in the phage and bacteria experiments. After the exposure, 100 μl of the yeast suspension was plated onto YPD agar plates. In each case, the plate count method was used to determine the viability of the yeasts. All experiments were performed in triplicate.4. UV-vis spectral analysis
UV-vis spectra were obtained using an Evolution 220 UV-visible spectrophotometer (Thermo Scientific, Waltham, Massachusetts, USA). All Congo red samples were diluted 100 times just before the measurement. Measurements were performed in quartz cuvettes (10 × 10 mm, Hellma). During all measurements, wavelengths from 200 nm to 800 nm were examined, with increments of 1 nm.5. The sterilization of Nylon66 membranes
To simulate the sterilization of the membranes in the bioreactors, 0.22 μm Nylon66 syringe filters were used. First, refreshed E. coli BL21 (DE3) GFP (OD600 = 1.23) or P. aeruginosa PAO1 (OD600 = 0.82) was diluted in 0.9% NaCl to obtain the concentration of bacteria about 104 CFU ml−1. The Nylon66 filters were flushed with 5 ml of bacterial cultures diluted in 0.9% NaCl using a syringe. The same sample was flushed through the filter twice. Then, the filters were filled with 1 ml of the following samples: 1) control (just TM buffer); 2) TM buffer and 15 minutes of UV exposure; 3) TM buffer and T4/LR1_PA01 phages (103 PFU ml−1); 4) TM buffer and Congo red; 5) TM buffer, T4/LR1_PA01, and UV; 6) TM buffer, CR and UV; 7) TM buffer, T4/LR1_PA01, and CR; 8) TM buffer, T4/LR1_PA01, CR, and UV. The final concentrations were around 103 PFU ml−1 of T4/LR1_PA01 phages and 0.1% CR. The samples were incubated for 15 minutes at room temperature and then exposed for 15 minutes to UV radiation within the laminar hood (Thermo Scientific MSC-ADVANTAGE; 36 W, 254 nm UV).To recover bacterial cells, the filters were washed with 1 mL of 0.9% NaCl solution. The number of bacteria was estimated by the plating method. 100 μl of each bacterial solution (diluted 1 and 10 times) was cultured overnight on LB-agar plates containing kanamycin (10 mg ml−1) at 37 °C. After the incubation, colonies were counted. For both control samples, a concentration of about 5 × 104 CFU (i.e., 5 ml of 104 CFU ml−1 suspension) was introduced, while approximately 4.8 × 104 CFU was recovered. Student's t-test was performed to evaluate whether observed differences were statistically significant (* p < 0.05; ** p < 0.01; *** p < 0.001). The experiment was conducted in triplicate.
Results & discussion
1. Bacteriophage, bacteria, and yeast exposure to UV
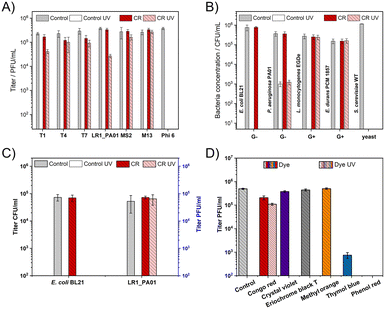
First, it was verified if 1 min of UV irradiation (254 nm, about 360 mJ cm−2) was enough to fully deactivate all studied bacteriophages. To cover various types of phages, T1, T4, T7, MS2, Phi 6, M13, and LR1_PA01 were studied. The energy used was similar to the case of the standard sterilization protocol.Bacteriophage Phi 6 was inactivated during the incubation with CR even before irradiation. When CR was added to the phage suspension, a minor (T4, T7) or no (T1, LR1_PA01, MS2, M13) decrease in the phage titer was observed upon 1 minute of UV irradiation. The decrease in the phage titer by about 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log for the LR1_PA01 phage was the biggest among all examined phages (Fig. 1A).
log for the LR1_PA01 phage was the biggest among all examined phages (Fig. 1A).
Gram-negative bacteria (E. coli BL21, P. aeruginosa PAO1), Gram-positive bacteria (L. monocytogenes EGDe, E. durans PCM 1857), and yeast (S. cerevisiae WT) were also a part of this study. CR alone did not affect the bacteria species (neither Gram-negative nor Gram-positive). P. aeruginosa appeared more resistant to UV compared to the other studied microorganisms. 1 minute UV irradiation caused only around 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log deactivation of P. aeruginosa compared to complete deactivation (5
log deactivation of P. aeruginosa compared to complete deactivation (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log to 6
log to 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log decrease) of E. coli, L. monocytogenes, E. durans, and S. cerevisiae. We also observed the elimination of S. cerevisiae caused by the incubation in CR, even without UV. The dye did not protect the Gram-negative bacteria against UV. The UV irradiation effect was very similar with and without CR. Meanwhile, CR provided almost complete protection against UV to the Gram-positive bacteria (both L. monocytogenes and E. durans) (Fig. 1B).
log decrease) of E. coli, L. monocytogenes, E. durans, and S. cerevisiae. We also observed the elimination of S. cerevisiae caused by the incubation in CR, even without UV. The dye did not protect the Gram-negative bacteria against UV. The UV irradiation effect was very similar with and without CR. Meanwhile, CR provided almost complete protection against UV to the Gram-positive bacteria (both L. monocytogenes and E. durans) (Fig. 1B).
In the next step of this experiment, the phages were exposed to UV radiation for an extended time (60 minutes). All non-enveloped phages (T1, T4, T7, LR1_PA01, MS2, M13) were protected against UV irradiation, yet the extent of protection varied slightly. In the case of M13, MS2, T4, and T7, the titer decreases were minimal. For T1, the decrease was around 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log, whereas for environmental phage LR1_PA01 it was around 1
log, whereas for environmental phage LR1_PA01 it was around 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log (ESI,† Fig. S1).
log (ESI,† Fig. S1).
To verify if the protective properties of CR depend on the phage titer, T1, T4, T7, LR1_PA01, MS2, and M13 at concentrations of 108, 107, and 106 PFU ml−1 were exposed to 1 minute of UV irradiation. Even at such high concentrations, 1 minute of exposure to UV was enough to inactivate non-protected phages to below the detection limits regardless of the initial concentration (up to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log). The protective properties of CR seem to be independent of the phage concentration. The survival rate in samples protected by CR was similar to the concentration of about 105 PFU ml−1 (ESI,† Fig. S2). That is, the decreases were up to about 1
log). The protective properties of CR seem to be independent of the phage concentration. The survival rate in samples protected by CR was similar to the concentration of about 105 PFU ml−1 (ESI,† Fig. S2). That is, the decreases were up to about 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log.
log.
Next, a mixture containing both the phage (LR1_PA01) and its non-host bacteria (E. coli) was prepared. This experiment was designed i) to prove the selectivity of CR towards phages and not bacteria and ii) to check if bacteria affected the phage-protective properties of CR. UV irradiation caused the complete deactivation of all bacteria cells (around 105 CFU ml−1 of E. coli) with and without CR. LR1_PA01 became deactivated by UV without CR but was fully protected in the presence of CR. The amount of active LR1_PA01 phages was approximately the same as before the irradiation (Fig. 1C).
Moreover, five more dyes were tested to verify the uniqueness of CR as a phage protectant. For this experiment, two other azo dyes (eriochrome black T and methyl orange) and three representatives of triphenylmethane dyes (crystal violet, thymol blue, and phenol red) were chosen (Fig. 1D). All of them are pH indicators used in cell biology laboratories. Titration tests revealed that only CR had bacteriophage protective properties against UV. In contrast, thymol blue and phenyl red caused the decrease of viral titers after 24 h incubation compared to the control sample and other tested compounds.
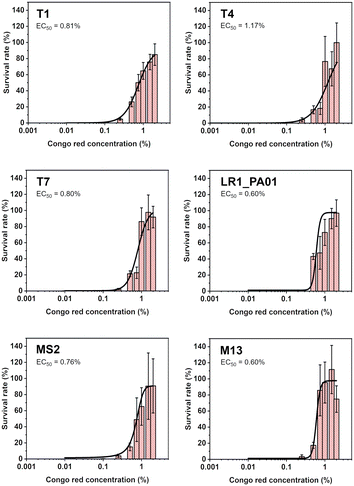
To verify the lowest effective dose of CR protecting against UV irradiation, bacteriophages were pre-incubated with the dye. The concentrations varied from 0.001% to 2%. The samples were exposed to UV radiation for 1 minute (the same parameters as in previous experiments). Data were fitted using the Hill equation to find EC50, i.e., the concentration that provides 50% protection. The EC50 was between 0.60% and 1.17% for all the phages (Fig. 2). The lowest concentration of CR, allowing any virions to survive, was about 0.1%.
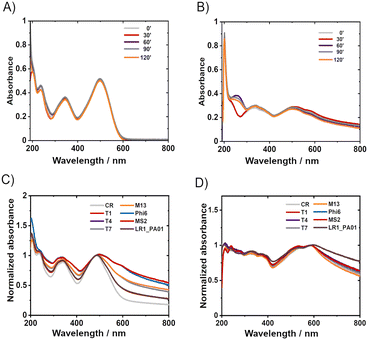
2. The mechanism of protection
UV-vis allowed for insight into the mechanisms of CR protective action. CR appeared stable in water after 24 hours of incubation or even after 120 min UV irradiation (Fig. 3A). Two leading bands in the spectra of CR in water, ca. 345 nm and 500 nm, correspond to π–π* transitions in the amino and azo groups, respectively.38 The absorbance of these bands did not significantly decrease upon UV irradiation (254 nm + 365 nm; 8 W).In PBS, the absorbance decreased slightly over time (ESI,† Fig. S5B), whereas UV irradiation didn't cause significant changes in the spectra (Fig. 3A). In the TM buffer, dramatic changes were observed within only 20 minutes, even without UV irradiation (ESI,† Fig. S5C). The absorbance decreased over 60 minutes, and the maximum shifted from 490 nm to about 580 nm while the spectrum changed its shape. Consecutive UV irradiation caused only slight changes (Fig. 3B). Partial precipitation of reddish sediment in the TM buffer occurred. Concentrated samples of CR in TM buffer (i.e., 0.5 mg ml−1) were turbid, in contrast to clear solutions of the same concentration in ultrapure water and PBS. The changes in the UV-vis spectra of Congo red in TM buffer upon storage (without UV irradiation) were due to the presence of divalent cations. The complexation of Mg2+ with CR explained the presence of the isosbestic point (∼580 nm). The further decrease of absorption with time, without changing the spectrum, probably corresponded to sedimentation, as described previously.39
TM was proved to have a significant impact on CR. Yet this could not be overcome by using different buffers (e.g., PBS) for phage-related experiments. It is proven that most phages require magnesium cations for appropriate infectivity.40 Phage DNA polymerases require divalent cations (Mg2+ or Mn2+) as the cofactor.41 Without Mg2+, the multiplication of phages is much slower than that under the same conditions (temperature and rate of infection, i.e., number of phages per single bacterial cell; ROI).42
Knowing the effects of the buffer constituents on CR, it was verified if the presence of bacteriophages caused changes in the UV-Vis spectra of Congo red. The measurements were conducted after 1 minute or 24 hours of incubation of phages in 1% Congo red solution in TM (Fig. 3C and D) and PBS buffers (ESI,† Fig. S5H and I). Phages alone in buffers (TM and PBS) absorb light only in a region below 250 nm.43 Hence phage capsids are exclusively made of proteins, and the proteins generally absorb wavelengths of about 230 nm and 260 nm.44 The bacteriophage spectra are not distinct; the absorption of radiation lies only in the range of 230–280 nm; this range is specific for the absorbance by proteins and nucleic acids.45 When phages were mixed with CR and incubated for 1 minute, a minor shift of the maximum CR absorption was observed from around 490 nm to 500 nm. Also, significant changes in the shape of the CR spectrum appeared in the region between 500 nm and 800 nm. A similar shift was observed for CR in protein suspension46 (ESI,† Fig. S7A and B) or in the presence of amyloids.47,48
CR–protein complexes are highly stable, and CR is believed to interact on multiple sides with proteins.49 These features suggest that CR interacts with phage virions by binding. The general shape of the spectrum was similar after 24 hours with and without phages, but the overall absorption intensity was much higher when phages were present. The fact that the shift was still present (although shifted to about 600 nm) suggested that the complex of CR and Mg2+ might be bound to phage protein capsids. We performed spectrophotometric titration and estimated the binding constant between CR and T4 virions to be around 1.28 × 104 dm3 mol−1 (cf. ESI†).
3. Membrane sterilization
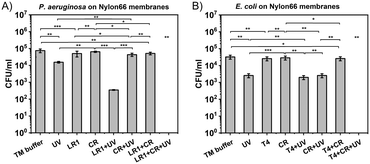
For the membrane sterilization protocol, the combination of bacteriophages (LR1_PA01 or T4), UV radiation, and Congo red was used. Nylon66 syringe filters were used as a model for sterilization of bioreactor membranes suffering from bacterial biofouling. The contamination of the membranes was simulated with P. aeruginosa PAO1 (Fig. 4A) or E. coli BL21 (DE3) GFP bacteria (Fig. 4B). First, the efficacy of only bacteriophages and only UV against biofouling was verified. Both factors caused a statistically significant decrease in bacteria by around 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log to 1
log to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log. Sterilization with only bacteriophages (from 7 × 104 to 5 × 104 CFU ml−1; from 3 × 104 to 2 × 104 CFU ml−1 for T4) was less effective than sterilization by UV (from 7 × 104 to 2 × 104 CFU ml−1; from 3 × 104 to 3 × 103 CFU ml−1). Next, we simultaneously used phages and UV. For T4, the effect was slightly better than using only UV (from 3 × 104 to 1.5 × 103 CFU ml−1). For LR1_PA01, the difference was much more significant (from 7 × 104 to 4 × 102 CFU ml−1). This was expected, as unprotected phages were deactivated by irradiation. Next, CR was introduced to the mix. There were no significant differences between UV and UV + CR, nor between phages and phages + CR. However, a combination of all three components, i.e., UV + phages + CR, resulted in the complete eradication of both E. coli and P. aeruginosa bacteria (i.e., from 3 × 104 CFU ml−1 to below the detection limits of the plating method ∼10 CFU ml−1). The total cleaning time was about 30 minutes, which provided added value to the UV- and phage-based sterilization protocol.
log. Sterilization with only bacteriophages (from 7 × 104 to 5 × 104 CFU ml−1; from 3 × 104 to 2 × 104 CFU ml−1 for T4) was less effective than sterilization by UV (from 7 × 104 to 2 × 104 CFU ml−1; from 3 × 104 to 3 × 103 CFU ml−1). Next, we simultaneously used phages and UV. For T4, the effect was slightly better than using only UV (from 3 × 104 to 1.5 × 103 CFU ml−1). For LR1_PA01, the difference was much more significant (from 7 × 104 to 4 × 102 CFU ml−1). This was expected, as unprotected phages were deactivated by irradiation. Next, CR was introduced to the mix. There were no significant differences between UV and UV + CR, nor between phages and phages + CR. However, a combination of all three components, i.e., UV + phages + CR, resulted in the complete eradication of both E. coli and P. aeruginosa bacteria (i.e., from 3 × 104 CFU ml−1 to below the detection limits of the plating method ∼10 CFU ml−1). The total cleaning time was about 30 minutes, which provided added value to the UV- and phage-based sterilization protocol.
4. Discussion
Phages were exposed to a dose of UV radiation of approximately 6 mJ cm−2 s−1, similar to the standard dose of energy used in laminar hoods for their sterilization. All studied phages were completely deactivated by 1 minute UV irradiation (around a 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log decrease in the number of active phages). Meanwhile, non-enveloped phages (T1, T4, T7, LR_PA01, MS2, M13) dispersed in 1% Congo red in TM buffer (EC50 values were between 0.60% and 1.17% for all phages) presented only a reasonable (no larger than one order of magnitude decrease) or no inactivation at all (Fig. 1). Remarkably, CR protected non-enveloped phages from much larger irradiation times – even after 60 minutes of irradiation, the titer decreases were only around 1
log decrease in the number of active phages). Meanwhile, non-enveloped phages (T1, T4, T7, LR_PA01, MS2, M13) dispersed in 1% Congo red in TM buffer (EC50 values were between 0.60% and 1.17% for all phages) presented only a reasonable (no larger than one order of magnitude decrease) or no inactivation at all (Fig. 1). Remarkably, CR protected non-enveloped phages from much larger irradiation times – even after 60 minutes of irradiation, the titer decreases were only around 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log (ESI,† Fig. S1). This protection appears regardless of the initial concentration of phages up to around 108 PFU ml−1 (ESI,† Fig. S2). The versatility of the developed approach was proved using the LR1_PA01 phage, i.e., an environmental isolate.
log (ESI,† Fig. S1). This protection appears regardless of the initial concentration of phages up to around 108 PFU ml−1 (ESI,† Fig. S2). The versatility of the developed approach was proved using the LR1_PA01 phage, i.e., an environmental isolate.
CR did not affect the fitness of bacteria (neither Gram-positive nor Gram-negative) but caused the complete eradication of yeast cells. This was in line with the paper by Suzuki et al., who proved the antifungal properties of Congo red. The authors showed that the dye binds to the beta-glucans in the chitin cell wall, prohibiting further chitin synthesis. This causes improper cell separation and the death of both mother and daughter cells.50,51
Two limitations of CR application were identified: CR had a negative impact on enveloped phages (in our case, Phi 6) and provided UV protection to Gram-positive bacteria. Both these limitations are not severe, as i) enveloped phages are relatively uncommon,52 and ii) Gram-negative strains that form biofilms are a much more significant problem for water-related research and MBR operation,53,54 compared to bacterial biofilms of Gram-positive bacteria (Enterococcus faecalis, Staphylococcus aureus, Viridans group Streptococci).55 Also, Gram-negative bacteria are much more commonly used in biotechnology and industry.56
Enveloped phage Phi 6 was deactivated by CR even without UV. CR is known to have detergent-like properties.57 Detergents were reported to destroy membranes and vesicle lipid structures.58 Destabilization of the phage envelope reduced the infectivity.59 This hypothesis was confirmed in our case by comparing the effect of 1% CR and 1% solution of a well-known surfactant – sodium dodecyl sulfate (SDS). SDS inactivated Phi 6 (enveloped phage) but not T4 (representative of non-enveloped phages) (ESI,† Fig. S3 and S4), exactly as CR did.
CR did not provide any protection to the Gram-negative bacteria against UV irradiation. However, the Gram-positive bacteria were fully protected. L. monocytogenes is well known for its resilience to adverse environmental conditions, including low temperatures,60 high concentrations of salt,61 and UV-C radiation.62 However, the survival of E. durans cells was most unexpected. According to the literature, for some bacteria, wall teichoic acids (WTAs) might be essential for the survival of the Gram-positive bacteria in the presence of Congo red.50 This report, along with our results, suggested that WTAs bind CR molecules at the surface of the bacterial cell, providing protection against UV.50
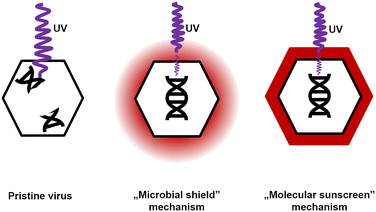
Two possible mechanisms explaining CR protection against UV were analyzed. The first scenario did not assume any specific interactions between CR and virions. The dye present in the solution absorbed some of the UV radiation. Bacteriophages and bacteria were still exposed, but the energy that reached them decreased. Because bacteriophages are much smaller than bacteria and yeasts, they absorb smaller, sublethal radiation doses. Hence, this mechanism was termed a ‘microbial shield’. The second scenario assumed the interaction between CR and the phage virions. The dye molecules bound to the surface of the virion absorbed UV radiation and dissipated the excess energy.63 CR present at the surface of the capsid absorbed a significant amount of radiation, so the viral genome was effectively exposed to smaller doses than objects without a CR protective layer. This mechanism was named a ‘molecular sunscreen’ (Fig. 5).
Evidence suggested that the ‘molecular sunscreen’ mechanism was a more plausible explanation.
i) Among tested dyes, only CR shows any protection against bacteriophages, even though almost all of these dyes absorb the radiation of the used wavelength, i.e., 254 nm (ESI,† Fig. S6A and B). Therefore, there should be some local increase in the concentration of CR, presumably due to interaction with protected entities, which was not present between virions and other studied dyes.
ii) The shifts in the UV-vis spectra of CR incubated with bacteriophages (225 → 230 nm and 490 → 500 nm) (Fig. 3C and D) were directly observed. Based on the literature47,48 and experimental results, this shift was interpreted as the binding of CR molecules to viral particles. The same shift was observed in CR solutions containing 10 μg ml−1 human serum albumin (ESI,† Fig. S7A and B). Also, similar shifts in the UV-vis spectra were reported for CR binding to β-amyloid fibers.38 For globular proteins (e.g., bovine serum albumin, similar to HSA), an additional shift was observed in the UV spectrum towards 225–230 nm.46,64
iii) CR deactivated Phi 6 (even without UV), thus proving that it indeed interacted with virions. Observations i) to iii) fit the results of Shapiro and co-workers,19 who described the protection against UV irradiation provided by CR to Lymantria dispar (L.) nuclear polyhedrosis virus (LdMNPV) (Baculoviridae, a group of viruses infecting insects).19 In this study, the authors observed a similar effect for another baculovirus – Heliothis zea (Boddie), a singly embedded nuclear polyhedrosis virus (HzSNPV).65 NPVs are different from ‘regular’ virions, as they are embedded within the protein polyhedrin matrix. It is most likely that Congo red bound on the surface of baculoviral occlusive bodies, locally accumulating CR molecules. This phenomenon might protect virions within the matrix. Eukaryotic viruses without such structures were unaffected or even deactivated by CR.34,35
iv) Gram-positive bacteria were protected by CR against UV, and Gram-negative bacteria were not. This was because of the accumulation of CR at the surface of Gram-positive bacteria due to interaction with cell wall components.50 This property is exploited in other bacteria-related studies; for instance, CR agar is routinely used to identify biofilm-forming bacterial strains, e.g., S. aureus.66
v) Spectrophotometric titration of CR solution with T4 bacteriophage suspension proved a correlation between the increase of absorbance (wavelength = 600 nm) and the increasing concentration of bacteriophages (ESI,† Fig. S8). The analysis of these results using Scatchard's method67 suggests that the complexation of CR with phage protein occurs in the tested range of bacteriophage concentration. The estimated value of the binding constant between CR and T4 phages is approximately 1.28 × 104 dm3 mol−1. This value is lower than the binding constant of CR and bovine serum albumin (BSA), i.e., 1.947 × 105 dm3 mol−1.64 The lower binding constant between CR and virions compared to CR and the model protein (BSA) was expected and might be explained by the varying amino acid composition of viral proteins or geometrical restrictions – in the experiments with free BSA, the complex formation is not hindered by any neighboring macromolecules (proteins or nucleic acids) as in the case of virions.
All these findings were used to fight biofouling. We tested 0.22 μm Nylon66 syringe filters contaminated with P. aeruginosa and E. coli (Fig. 4A and B, ESI,† Fig. S9) as a simplified model of bioreactor membrane biofouling. The efficacy of bacteriophages, UV, CR, and their combinations against bacteria embedded on the membrane surface was examined in this design. Depending on bacterial species, differences in the efficiency of examined factors may occur, e.g., E. coli was more sensitive to just UV exposure, while P. aeruginosa presented increased sensitivity to the combination of phages and UV. Nevertheless, in this study, combining all three factors of the ‘cleaning mixture’, i.e., a combination of phages, UV radiation, and Congo red, was effective for membrane sterilization from both bacterial contaminations within just 30 minutes. However, in a complex system like biofilms, as presented by Scarascia et al.,10 the total time of sterilization and its efficacy might differ due to the differences in laboratory-scale methods and their field applications.68
The proposed method for membrane sterilization doesn't affect the integrity of the membrane, which was proven with optical microscopy examination (ESI,† Fig. S10). The remains of the dye after the cleaning procedure can be efficiently removed from the membrane by rinsing it with water. Estimation suggests that about 4 liters of water per m2 of the membrane, divided for six rinses, would be enough for removing the remains of CR to the values undetectable with spectroscopic techniques (ESI,† Fig. S11 and S12).
The ‘cleaning mixture’ reusability was also proven for the T4 phage. Hence the T4 phage is a representative of the most widely distributed (and frequently used) type of bacteriophage belonging to the Caudovirales order. It is most probably the ‘cleaning mixture’ containing other phages that could also be reused. The sterilization efficiency drops significantly after the second cycle (ESI,† Fig. S13). Nevertheless, this clear advantage saves money (estimated costs of simultaneous sterilization with bacteriophages and UV is about 50% lower compared to the sterilization with just UV, see the ESI†), operation time, and labor. This allows using the same ‘cleaning mixture’ to disinfect tandem operational units in larger foundries.
Conclusions
The combination of phages, Congo red, and UV radiation was successfully applied to sterilize membrane filters simulating bioreactors. The addition of CR allowed for the simultaneous action of UV and phages. The dye acted as a ‘molecular sunscreen’, protecting phages but not Gram-negative bacteria from harmful irradiation. Its antifungal properties against yeasts provide additional value for sterilization purposes. All these findings allowed us to propose the sterilization protocol requiring only about 30 minutes. The method is adequate for non-enveloped phages and Gram-negative bacteria. The additional component, i.e., CR, is cheap and has relatively low cytotoxicity (comparable to caffeine). Moreover, the ‘cleaning mixture’ can be reused, and ultimately CR can be removed from the aqueous solutions using the previously described protocol.69–71 This would potentially allow limiting the use of chemicals for bioreactor membrane sterilization and, therefore, the risk of the release of chemicals into the environment.The presented study is the first attempt to investigate the effect of Congo red on bacteriophages. CR protected non-enveloped phages (including environmentally isolated phages) but deactivated the enveloped Phi 6 phage. For the explanation of the CR effect on different viruses, the ‘molecular sunscreen’ mechanism, i.e., CR bound to virions providing local protection, was proposed and confirmed during the experiments. There were no specific interactions between CR and Gram-negative bacteria, which remained unprotected and vulnerable to both UV and phages.
Author contributions
M. W.: conceptualization, writing the first draft, original draft preparation, editing; P. M.: conceptualization, writing the first draft; R. Z.: original draft preparation, editing; B. B.: editing; J. P.: conceptualization, original draft preparation, supervision, investigation, writing – review, and project administration.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was financed by the National Science Centre within the SONATA BIS grant according to decision number 2017/26/E/ST4/00041. M. W. was partially financed by the Foundation for Polish Science from the European Regional Development Fund within the project POIR.04.04.00-00-14D6/18-00 ‘Hybrid sensor platforms for integrated photonic systems based on ceramic and polymer materials (HYPHa)’ (TEAM-NET program).References
- Markets and Markets, Market Research Report, 2022.
- Markets and Markets, Membrane Bioreactor Market, 2020.
- Membrane Bioreactors: Global Markets.
- N. Porcelli and S. Judd, Chemical cleaning of potable water membranes: A review, Sep. Purif. Technol., 2010, 71, 137–143 CrossRef CAS.
- K. Kimura, Y. Hane, Y. Watanabe, G. Amy and N. Ohkuma, Irreversible membrane fouling during ultrafiltration of surface water, Water Res., 2004, 38, 3431–3441 CrossRef CAS PubMed.
- Z. Atamer, S. Meike, H. Neve, K. J. Heller and J. Hinrichs, Review: Elimination of bacteriophages in whey and whey products, Front. Microbiol., 2013, 4, 1–9 Search PubMed.
- M. Berovic, Sterilisation in biotechnology, Biotechnol. Annu. Rev., 2005, 11, 257–279 CAS.
- G. U. Semblante, F. I. Hai, D. D. Dionysiou, K. Fukushi, W. E. Price and L. D. Nghiem, Holistic sludge management through ozonation: A critical review, J. Environ. Manage., 2017, 185, 79–95 CrossRef CAS PubMed.
- Y. N. Chang and F. I. Wei, High-temperature chlorine corrosion of metals and alloys - A review, J. Mater. Sci., 1991, 26, 3693–3698 CrossRef CAS.
- G. Scarascia, L. Fortunato, Y. Myshkevych, H. Cheng, T. O. Leiknes and P. Y. Hong, UV and bacteriophages as a chemical-free approach for cleaning membranes from anaerobic bioreactors, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, 1–9 CrossRef PubMed.
- E. A. Wójcik, M. Stanczyk, A. Wojtasik, J. D. Kowalska, M. Nowakowska, M. Lukasiak, M. Bartnicka, J. Kazimierczak and J. Dastych, Comprehensive Evaluation of the Safety and Efficacy of BAFASAL® Bacteriophage Preparation for the Reduction of Salmonella in the Food Chain, Viruses, 2020, 12(7), 742 CrossRef PubMed.
- J. Baj, Ł. Dziewit, D. Bartosik, M. Radlińska, B. Paterczyk and Ł. Drewniak, Mikrobiologia, PWN, Warszawa, 2018 Search PubMed.
- D. Trasanidou, A. S. Gerós, P. Mohanraju, A. C. Nieuwenweg, F. L. Nobrega and R. H. J. Staals, Keeping crispr in check: Diverse mechanisms of phage-encoded anti-crisprs, FEMS Microbiol. Lett., 2019, 366, 1–14 CrossRef PubMed.
- CDC, Biggest Threats and Data 2019 AR Threats Report.
- Acute Market Reports, Bacteriophage Market, 2021.
- B. Wu, R. Wang and A. G. Fane, The roles of bacteriophages in membrane-based water and wastewater treatment processes: A review, Water Res., 2017, 110, 120–132 CrossRef CAS PubMed.
- M. Ben Said, O. Masahiro and A. Hassen, Detection of viable but non cultivable Escherichia coli after UV irradiation using a lytic Qβ phage, Ann. Microbiol., 2010, 60, 121–127 CrossRef CAS PubMed.
- M. Ben Said, M. Otaki and A. Hassen, Use of lytic phage to control Salmonella typhi's viability after irradiation by pulsed UV light, Ann. Microbiol., 2012, 62, 107–111 CrossRef.
- M. Shapiro, Congo Red as an Ultraviolet Protectant for the Gypsy Moth (Lepidoptera: Lymantriidae) Nuclear Polyhedrosis Virus, J. Econ. Entomol., 1989, 82, 548–550 CrossRef.
- R. Sommer, W. Pribil, S. Appelt, P. Gehringer, H. Eschweiler, H. Leth, A. Cabaj and T. Haider, Inactivation of bacteriophages in water by means of non-ionizing (UV-253.7nm) and ionizing (gamma) radiation: A comparative approach, Water Res., 2001, 35, 3109–3116 CrossRef CAS PubMed.
- M. Norval and A. A. El-Ghorr, UV radiation and mouse models of herpes simplex virus infection, Photochem. Photobiol., 1996, 64, 242–245 CrossRef CAS PubMed.
- Q. S. Meng and C. P. Gerba, Comparative inactivation of enteric adenoviruses, poliovirus and coliphages by ultraviolet irradiation, Water Res., 1996, 30, 2665–2668 CrossRef CAS.
- E. F. Tom, I. J. Molineux, M. L. Paff and J. J. Bull, Experimental evolution of UV resistance in a phage, PeerJ, 2018, 2018, 1–20 Search PubMed.
- D. Ma, B. Gao, D. Hou, Y. Wang, Q. Yue and Q. Li, Evaluation of a submerged membrane bioreactor (SMBR) coupled with chlorine disinfection for municipal wastewater treatment and reuse, Desalination, 2013, 313, 134–139 CrossRef CAS.
- S. Tang, J. Li, Z. Zhang, B. Ren and X. Zhang, Comparison of long-term ceramic membrane bioreactors without and with in-situ ozonation in wastewater treatment: Membrane fouling, effluent quality and microbial community, Sci. Total Environ., 2019, 652, 788–799 CrossRef PubMed.
- M. Połaska and B. Sokołowska, Review bacteriophages—a new hope or a huge problem in the food industry, AIMS Microbiol., 2019, 5, 324–347 Search PubMed.
- P. Frid, S. V. Anisimov and N. Popovic, Congo red and protein aggregation in neurodegenerative diseases, Brain Res. Rev., 2007, 53, 135–160 CrossRef CAS PubMed.
- J. L. Wang, L. Zhao, M. Q. Li, W. G. Chen and C. J. Xu, A sensitive and reversible staining of proteins on blot membranes, Anal. Biochem., 2020, 592, 113579 CrossRef CAS PubMed.
- J. L. Wang, M. Q. Li, J. J. Zhang and C. J. Xu, Total protein staining with Congo red as an alternative loading control for western blot analysis, Biotech. Histochem., 2022, 97, 404–414 CrossRef CAS PubMed.
- R. Khurana, V. N. Uversky, L. Nielsen and A. L. Fink, Is Congo Red an Amyloid-specific Dye?, J. Biol. Chem., 2001, 276, 22715–22721 CrossRef CAS PubMed.
- M. Tubis, W. H. Blahd and R. A. Nordyke, The preparation and use of radioiodinated Congo red in detecting amyloidosis, J. Am. Pharm. Assoc., Sci. Ed., 1960, 49, 422–425 CrossRef CAS.
- J. M. Peters, Factors Affecting Caffeine Toxicity: A Review of the Literature, J. Clin. Pharmacol. J. New Drugs, 1967, 7, 131–141 CrossRef CAS.
- B. D. Rawal and G. N. Vyas, Magnesium-mediated reversal of the apparent virucidal effect of ascorbic acid or congo red reacted in vitro with the human immunodeficiency virus, Biologicals, 1996, 24, 113–116 CrossRef CAS PubMed.
- J. Estupinan and R. P. Hanson, Congo red and trypan blue as stains for plaque assay of newcastle disease virus, Avian Dis., 1969, 13, 330–339 CrossRef CAS PubMed.
- H. Becht and R. Drzeniek, The effect of azo dyes on myxovirus neuraminidase and on virus multiplication, J. Gen. Virol., 1968, 2, 261–268 CrossRef CAS PubMed.
- K. Scott, R. E. Kerber and A. Mügge, Free Radicals From Plastic Syringes, Free Radical Biol. Med., 1991, 11, 69–70 CrossRef PubMed.
- Ł. Richter, K. Księżarczyk, K. Paszkowska, M. Janczuk-Richter, J. Niedziółka-Jönsson, J. Gapiński, M. Łoś, R. Hołyst and J. Paczesny, Adsorption of bacteriophages on polypropylene labware affects the reproducibility of phage research, Sci. Rep., 2021, 11, 1–12 CrossRef PubMed.
- K. Yokoyama, A. D. Fisher, A. R. Amori, D. R. Welchons and R. E. McKnight, Spectroscopic and calorimetric studies of congo red dye-amyloid peptide complexes, J. Biophys. Chem., 2010, 01, 153–163 CrossRef CAS.
- N. J. Ellis and D. M. Bishop, Congo Red As an Indicator for Magnesium. I. a Comparison of Serum, Can. J. Biochem. Physiol., 1964, 42, 1225–1231 CrossRef CAS PubMed.
- L. Yang, K. Arora, W. A. Beard, S. H. Wilson and T. Schlick, Critical role of magnesium ions in DNA polymerase β's closing and active site assembly, J. Am. Chem. Soc., 2004, 126, 8441–8453 CrossRef CAS PubMed.
- S. Tabor and C. C. Richardson, Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 4076–4080 CrossRef CAS PubMed.
- R. G. Tucker, The Role of Magnesium Ions in the Growth of Salmonella Phage Anti-R, Microbiology, 1961, 26(2), 313–323 CrossRef CAS PubMed.
- A. Bérces, M. Egyeki, A. Fekete, G. Horneck, G. Kovács, C. Panitz and G. Rontó, The PUR experiment on the EXPOSE-R facility: Biological dosimetry of solar extraterrestrial UV radiation, Int. J. Astrobiol., 2015, 14, 47–53 CrossRef.
- R. Stefanescu, S. Brebu, M. Matei, I. M. Risca, A. Surleva and G. Drochioiu, Contribution to Casein Determination by UV Spectrophotometry, Acta Chem. Iasi, 2017, 25, 112–126 CrossRef.
- J. R. Whitaker and P. E. Granum, An absolute method for protein determination based on difference in absorbance at 235 and 280 nm, Anal. Biochem., 1980, 109, 156–159 CrossRef CAS PubMed.
- D. N. De Vasconcelos and V. F. Ximenes, Albumin-induced circular dichroism in Congo red: Applications for studies of amyloid-like fibril aggregates and binding sites, Spectrochim. Acta, Part A, 2015, 150, 321–330 CrossRef PubMed.
- T. Miura, C. Yamamiya, M. Sasaki, K. Suzuki and H. Takeuchi, Binding mode of Congo Red to Alzheimer's amyloid β-peptide studied by UV Raman spectroscopy, J. Raman Spectrosc., 2002, 33, 530–535 CrossRef CAS.
- W. E. Klunk, R. F. Jacob and R. P. Mason, Quantifying amyloid by congo red spectral shift assay, Methods Enzymol., 1999, 309, 285–305 CAS.
- H. M. C. de Paula, Y. L. Coelho, A. J. P. Agudelo, J. P. de Rezende, G. M. D. Ferreira, G. M. D. Ferreira, A. C. S. dos Pires and L. H. M. da Silva, Kinetics and thermodynamics of bovine serum albumin interactions with Congo red dye, Colloids Surf., B, 2017, 159, 737–742 CrossRef PubMed.
- T. Suzuki, J. Campbell, Y. Kim, J. G. Swoboda, E. Mylonakis, S. Walker and M. S. Gilmore, Wall teichoic acid protects Staphylococcus aureus from inhibition by Congo red and other dyes, J. Antimicrob. Chemother., 2012, 67, 2143–2151 CrossRef CAS PubMed.
- M. Kopecká and M. Gabriel, The influence of Congo red on the cell wall and (1 → 3)-β-d-glucan microfibril biogenesis in Saccharomyces cerevisiae, Arch. Microbiol., 1992, 158, 115–126 CrossRef PubMed.
- M. M. Poranen and S. Mäntynen, ICTV virus taxonomy profile: Cystoviridae, J. Gen. Virol., 2017, 98, 2423–2424 CrossRef CAS PubMed.
- S. Ayyaru, J. Choi and Y. H. Ahn, Biofouling reduction in a MBR by the application of a lytic phage on a modified nanocomposite membrane, Environ. Sci.: Water Res. Technol., 2018, 4, 1624–1638 RSC.
- A. Lee, J. W. Elam and S. B. Darling, Membrane materials for water purification: Design, development, and application, Environ. Sci.: Water Res. Technol., 2016, 2, 17–42 RSC.
- Z. Khatoon, C. D. McTiernan, E. J. Suuronen, T. F. Mah and E. I. Alarcon, Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention, Heliyon, 2018, 4, e01067 CrossRef PubMed.
- W. M. De Vos, M. Kleerebezem and O. P. Kuipers, Expression systems for industrial Gram-positive bacteria with low guanine and cytosine content, Curr. Opin. Biotechnol., 1997, 8, 547–553 CrossRef CAS PubMed.
- C. Lendel, B. Bolognesi, A. Wahlström, C. M. Dobson and A. Gräslund, Detergent-like interaction of congo red with the amyloid β peptide, Biochemistry, 2010, 49, 1358–1360 CrossRef CAS PubMed.
- J. R. Henriksen, T. L. Andresen, L. N. Feldborg, L. Duelund and J. H. Ipsen, Understanding detergent effects on lipid membranes: A model study of lysolipids, Biophys. J., 2010, 98, 2199–2205 CrossRef CAS PubMed.
- S. Watts, M. Ramstedt and S. Salentinig, Ethanol Inactivation of Enveloped Viruses: Structural and Surface Chemistry Insights into Phi6, J. Phys. Chem. Lett., 2021, 12, 9557–9563 CrossRef CAS PubMed.
- S. Liu, J. E. Graham, L. Bigelow, P. D. Morse and B. J. Wilkinson, Identification of Listeria monocytogenes genes expressed in response to growth at low temperature, Appl. Environ. Microbiol., 2002, 68, 1697–1705 CrossRef CAS PubMed.
- O. Duché, F. Trémoulet, P. Glaser and J. Labadie, Salt stress proteins induced in Listeria monocytogenes, Appl. Environ. Microbiol., 2002, 68, 1491–1498 CrossRef PubMed.
- E. Gayán, M. J. Serrano, R. Pagán, I. Álvarez and S. Condón, Environmental and biological factors influencing the UV-C resistance of Listeria monocytogenes, Food Microbiol., 2015, 46, 246–253 CrossRef PubMed.
- A. L. Costa, A. C. Gomes, M. Pillinger, I. S. Gonçalves, J. Pina and J. S. Seixas de Melo, Insights into the Photophysics and Supramolecular Organization of Congo Red in Solution and the Solid State, ChemPhysChem, 2017, 18, 564–575 CrossRef CAS PubMed.
- Y. Z. Zhang, X. Xiang, P. Mei, J. Dai, L. L. Zhang and Y. Liu, Spectroscopic studies on the interaction of Congo Red with bovine serum albumin, Spectrochim. Acta, Part A, 2009, 72, 907–914 CrossRef PubMed.
- C. M. Ignoffo, B. S. Shasha and M. Shapiro, Sunlight ultraviolet protection of the Heliothis nuclear polyhedrosis virus through starch-encapsulation technology, J. Invertebr. Pathol., 1991, 57, 134–136 CrossRef.
- J. S. Lee, Y. M. Bae, A. Han and S. Y. Lee, Development of Congo red broth method for the detection of biofilm-forming or slime-producing Staphylococcus sp, LWT--Food Sci. Technol., 2016, 73, 707–714 CrossRef CAS.
- G. Scatchard, Statistical mechanics and reaction rates in liquid solutions, Chem. Rev., 1932, 10, 229–240 CrossRef CAS.
- S. Lee, S. K. Park, H. Kwon, S. H. Lee, K. Lee, C. H. Nahm, S. J. Jo, H. S. Oh, P. K. Park, K. H. Choo, C. H. Lee and T. Yi, Crossing the Border between Laboratory and Field: Bacterial Quorum Quenching for Anti-Biofouling Strategy in an MBR, Environ. Sci. Technol., 2016, 50, 1788–1795 CrossRef CAS PubMed.
- S. Radoor, J. Karayil, J. Parameswaranpillai and S. Siengchin, Removal of anionic dye Congo red from aqueous environment using polyvinyl alcohol/sodium alginate/ZSM-5 zeolite membrane, Sci. Rep., 2020, 10, 1–15 CrossRef PubMed.
- M. K. Purkait, A. Maiti, S. DasGupta and S. De, Removal of congo red using activated carbon and its regeneration, J. Hazard. Mater., 2007, 145, 287–295 CrossRef CAS PubMed.
- N. H. H. Hairom, A. W. Mohammad and A. A. H. Kadhum, Nanofiltration of hazardous Congo red dye: Performance and flux decline analysis, J. Water Process. Eng., 2014, 4, 99–106 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ew00913g |
| This journal is © The Royal Society of Chemistry 2023 |