Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Stay in the loop: lessons learned about the microbial water quality in pipe loops transitioned from conventional to direct potable reuse water†
Lauren C.
Kennedy‡
 abc,
Scott E.
Miller‡
abc,
Scott E.
Miller‡
 ab,
Rose S.
Kantor
ab,
Hannah
Greenwald
ab,
Rose S.
Kantor
ab,
Hannah
Greenwald
 ab,
Michael J.
Adelman
d,
Hari
Seshan
e,
Paige
Russell
ab,
Michael J.
Adelman
d,
Hari
Seshan
e,
Paige
Russell
 f and
Kara L.
Nelson
f and
Kara L.
Nelson
 *ab
*ab
aDepartment of Civil and Environmental Engineering, College of Engineering, University of California, Berkeley, CA, USA. E-mail: karanelson@berkeley.edu
bNational Science Foundation Engineering Research Center for Re-inventing the Nation's Urban Water Infrastructure, Berkeley, CA, USA
cDepartment of Civil and Environmental Engineering, College of Engineering, Stanford University, Stanford, CA, USA
dStantec, Pasadena, CA, USA
eTyr Group, Brisbane, Queensland, Australia
fBrown and Caldwell, San Diego, CA, USA
First published on 8th March 2023
Abstract
Direct potable reuse offers water-scarce regions a local and reliable source of drinking water, but few studies have investigated the effect of introducing advanced-treated wastewater to drinking water distribution systems on microbial water quality. We aimed to characterize the impact on microbial water quality of transitioning a drinking water distribution system fed with conventionally-treated surface water to a blend with advanced-treated wastewater. Three pipe loops were fed with the same source water (conventional water alone or blended with advanced-treated wastewater) for 21 weeks, and two pipe loops transitioned from conventional water to advanced-treated wastewater (i.e., conditioned reverse osmosis permeate) after the first 10 weeks. Microbial water quality was evaluated via flow cytometry-based cell counts, adenosine triphosphate concentrations, quantitative PCR, and 16S rRNA gene amplicon sequencing. Handling (i.e., conveyance, transportation, and storage) of reverse osmosis permeate introduced bioavailable carbon, which resulted in high microbial abundance in the advanced-treated wastewater that was fed to the pipe loops. In the pipe loops, the microbial community profile in the bulk water and biofilm reflected that of the primary feedwater (i.e., conventionally-treated surface water or advanced-treated wastewater). The antibiotic resistance gene sul1 was detected in all samples, increased in absolute abundance in the RO permeate after handling, and increased in relative and absolute abundance in the bulk water and biofilm of pipe loops after the transition. In addition, 21 amplicon sequence variants (ASVs) were significantly enriched or depleted in the bulk water and biofilm of pipe loops after the transition Of these ASVs, one Pseudomonas-classified ASV was present in the RO permeate, increased in estimated absolute abundance during RO handling, and remained at high estimated absolute abundance in pipe loops fed with primarily advanced-treated wastewater. Thus, advanced-treated wastewater has potential to be a source of antibiotic resistance and opportunistic pathogens in drinking water distribution systems. Though these contaminants will likely be low in abundance following treatment, nutrients introduced during storage or conveyance of advanced-treated wastewater could increase the abundance of antibiotic resistance genes and opportunistic pathogens. To ensure that risks are not elevated compared to conventional source waters, enhanced monitoring is recommended, that begins two years before the transition to direct potable reuse, continues for two years after, and includes absolute microbial abundance as well as screening for opportunistic pathogens and antibiotic resistance genes.
Water impactDirect potable reuse is becoming a realistic option for water-stressed cities, but there are knowledge gaps about the microbial impacts from introducing advanced-treated wastewater to drinking water distribution systems. Engineering consultants, water utility representatives, and academic researchers collaborated on this research that will inform future bench- and pilot-scale studies as well as full-scale direct potable reuse systems. |
1. Introduction
Direct potable reuse offers water-scarce regions a local and reliable source of drinking water through advanced treatment of wastewater. A growing body of research has demonstrated that advanced treatment of wastewater can yield a finished water that meets or exceeds water quality standards for drinking water by providing treatment barriers against pathogens and chemical contaminants.1–5 Direct potable reuse alternatives include i) raw water augmentation, in which advanced-treated wastewater is blended with raw water supplies upstream of a drinking water treatment plant and ii) treated water augmentation, in which advanced-treated wastewater is directly introduced into a drinking water distribution system. In some cases, direct potable reuse is an attractive alternative for reuse systems because it could reduce costs and energy associated with conveyance, pumping, and storage in environmental buffers.6 Potable reuse in general has system-level impacts on all aspects of the engineered water cycle, and direct potable reuse in particular has important water quality implications for drinking water systems.7 However, few studies have investigated the impacts that direct potable reuse may have on chemical and microbial water quality in drinking water distribution systems and premise plumbing.8–11 One recent bench-scale study investigated the effects of DPR blending on treatability at a conventional drinking water plant, but noted potential impacts on distribution system water chemistry and microbiology as topics for future study.12Drinking water distribution system water quality is a topic of great importance, with particular interest in improving the fundamental understanding of microbial water quality and biofilms. Drinking water distribution systems contain biofilm microbial communities, and changes in feedwater quality in the system have been shown to affect biofilm microbial community composition and absolute abundance.13–15 The water quality of advanced-treated wastewater can differ from conventional drinking water in ways that could affect the microbial water quality of the drinking water distribution system. For example, advanced treatment using reverse osmosis or nanofiltration will yield permeate with low concentrations of organic carbon,13,16 which could affect raw or treated water augmentation direct potable reuse systems. Based on studies of conventional drinking water distribution systems, lower concentrations of carbon, nutrients, or ions in advanced-treated wastewater can lead to sloughing of existing loose deposits and biofilm,14,17 reduced rates of chlorine decay during distribution,18 and reduced growth of distribution system microorganisms13 (e.g., opportunistic pathogens such as Mycobacterium spp. and Legionella pneumophila). For example, previous studies have observed significant, positive correlations between organic carbon and Mycobacterium spp.,19Legionella pneumophila,20 and Vermamoeba vermiformis20 (formerly known as Hartmanella vermiformia; an amoebal host for Legionella pneumophila21) in full-scale and simulated distribution systems.
Direct potable reuse, and particularly treated water augmentation, is still an emerging approach to potable reuse. Raw water augmentation projects and the associated regulatory framework are underway in several regions, and, to date, only three full-scale treated water augmentation systems are documented in the world.22 Because these systems are uncommon, bench- and pilot-scale simulation experiments can help identify microbial issues that could arise prior to new full-scale implementation. However, simulating a drinking water distribution system effectively is difficult and care must be taken in extending the results to full-scale systems. In one study simulating an upgrade of a conventional drinking water treatment facility to include membrane filtration, organic carbon and biomass decreased in bulk water and biofilm.13,16 However, full-scale upgrades in conventional treatment have been associated with immediate increases in biofilm sloughing.14,23 One reason for discrepancies between simulated and full-scale system findings is that tradeoffs must be made during the design of simulated systems. In particular, decisions are made about which design parameters or operational conditions are prioritized to mimic a full-scale system (e.g., shear force, flow rate, or water age).24 Studies of microbial water quality in particular require additional design considerations, such as including a sufficient conditioning period to establish biofilm in the simulated distribution system,25–28 obtaining samples with enough biomass for analyses, and achieving realistic primary and secondary disinfectant conditions. While there are plans for both raw and treated water augmentation systems (e.g., in San Diego, CA and El Paso, TX1 respectively), there are major knowledge gaps regarding the microbial impacts of a transition from an existing conventional system to one that incorporates direct potable reuse.8,10,9,11 Another challenge is that studying these impacts requires co-location of advanced-treated wastewater and conventional drinking water. Given the rarity of full-scale or treated water augmentation systems as well as the importance of filling knowledge gaps about these systems, more guidance is needed for simulated treated water augmentation system study design.
The objectives of this work were to: (i) characterize the impact on microbial water quality from transitioning a pilot-scale piped drinking water distribution system fed with conventionally-treated surface water to a treated water augmentation system; and (ii) evaluate the tradeoffs made in the study design to provide recommendations for future direct potable reuse simulation studies. We partnered with a water utility to design and operate an experimental setup of five simulated distribution systems (i.e., pipe loops). The pipe loops were fed different blends of conventionally-treated and advanced-treated waters over the course of two phases of operation (in total 21 weeks; Fig. 1), during which bulk water and biofilm samples were collected from each pipe loop to assess microbial water quality. Microbial water quality was evaluated via flow cytometry-based cell counts, adenosine triphosphate (ATP) concentration, quantitative PCR (qPCR), and 16S rRNA gene amplicon sequencing. qPCR targets were the 16S rRNA gene, one free-living amoeba, two opportunistic pathogens, and two antibiotic resistance genes. To our knowledge, this study is the first to investigate the microbial impacts of the transition from a conventional drinking water distribution system to a treated water augmentation system.
2. Methods
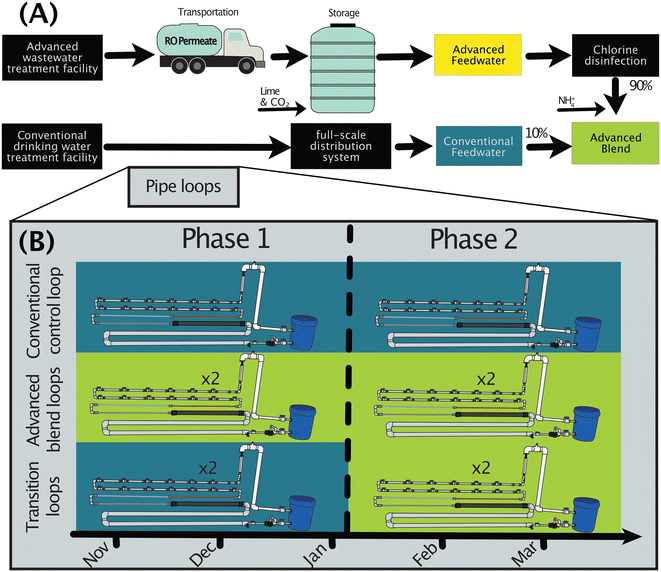
To simulate a transition of a drinking water distribution system to treated water augmentation, conventionally-treated drinking water and advanced-treated wastewater were fed to five pilot-scale pipe loops in a covered, outdoor area at a conventional drinking water treatment facility (Fig. 1).2.1. Origins of pipe loop feedwaters
The chloraminated conventionally-treated drinking water was obtained onsite (“conventional feedwater”). The advanced-treated wastewater (“advanced feedwater”) originated from a demonstration-scale advanced wastewater treatment facility to demonstrate suitability for potable reuse, at which tertiary-treated wastewater effluent underwent ozonation, biologically active carbon filtration, microfiltration, and reverse osmosis (“RO”) before collection for this study. RO permeate was transported via tanker truck from the advanced wastewater treatment facility and stored in a tank onsite at the drinking water treatment facility until use as advanced feedwater (Fig. 1A). Post-RO stabilization is an important step in full-scale potable reuse systems, so the advanced feedwater was conditioned with calcium hydroxide (“lime”) and carbon dioxide gas to achieve an alkalinity of 75–100 mg L−1 as CaCO3 and a pH of 8 in the range of typical post-stabilization targets. To compare the water quality in the advanced feedwater to that of the RO permeate, we utilized a previously published dataset for the RO permeate.29To compare the water quality in the pipe loops to that in the full-scale distribution system, three additional sites in the distribution system were sampled and combined with a previously published dataset on microbial abundance (DWDS F)30 from the same system (“full-scale distribution system”). Utility drinking water distribution system taps were sampled six times between February 14, 2018 and April 26, 2018, and the sites were selected to include a range in water age (50.6 h, 70.0 h, and 289 h). The water ages were provided by the utility based on an internal model (SynerGEE Water; v4.7.0).
2.2. Pipe loop design and start-up
To mimic the materials used in the full-scale distribution system and premise plumbing, the five identical pipe loops were constructed using copper, galvanized iron, leaded brass, and cement-lined ductile iron pipe segments, with the majority of piping made from PVC (Fig. S1†). Twelve removable segments of 0.3 m length PVC pipe, with true-union fittings and isolation valves between each segment, were installed in each pipe loop for biofilm collection. Pipe lengths and diameters are provided in the ESI.†Each loop included a reservoir for blending, a recirculation pump, a sample tap, a rotameter, and a hydraulic loop to ensure full flow in all pipes (Fig. S1†). The hydraulic loop had a syringe filter on its air vent to prevent intrusion of airborne bacteria. Water was recirculated continuously in each pipe loop at a flow rate of 30.3 L min−1 (8 gallons per minute). This achieved a fluid shear of 0.25 N m−2 on the inner surfaces of each PVC pipe segment used for biofilm collection, to simulate typical shear forces in drinking water distribution pipes.24 The pipe loops were designed to recirculate 100 L batches of water with residence times roughly equal for both the piping and reservoir sections of each pipe loop (1.9 and 1.8 minutes, respectively).
During start up, pipe loops were disinfected by recirculating 100 L of conventional drinking water with a high free chlorine residual (100 mg L−1 as Cl2), rinsed with four full batches of conventional drinking water, and then inoculated with concentrated biomass from the full-scale drinking water distribution system. The concentrated inoculum biomass was divided evenly across the five pipe loops and mixed with a fresh 100 L batch of conventional feedwater. The pipe loops recirculated the inoculum for seven days, after which, Phase 1 of the study began (Fig. 1B). More information on pipe loop start up can be found in the ESI† (e.g., inoculum preparation, inoculation of the pipe loops).
2.3. Pipe loop batching and operation
The pipe loops recirculated conventional feedwater that was either unaltered (“conventional”) or blended with advanced-treated wastewater (“advanced blend”) (Fig. 1B). The advanced blend contained 10% conventional feedwater and 90% advanced feedwater. The partner utility expected to have an annual average of 40% advanced-treated wastewater in the planned full-scale system. However, 90% advanced-treated wastewater was selected as a conservative but realistic blending scenario (e.g., drought conditions). Two pipe loops were fed with the advanced blend throughout the study (“advanced blend loops”), two were fed with conventional feedwater for 10 weeks followed by the advanced blend for 11 weeks (“transition loops”), and one was fed with conventional feedwater throughout the study (“conventional control loop”). The pipe loops were operated in semi-batch mode such that the loops were fully drained and replenished with fresh feedwater every 3.5 days, which simulated a conservative water age for the full-scale drinking water distribution system (only 12% of the network contains water with an age of three days or more).To prepare a batch of water for recirculation, 100 L of conventional feedwater was added into the pipe loop reservoir for pipe loops fed with conventional feedwater. For pipe loops fed with the advanced blend (i.e., the advanced blend loops in Phases 1 and 2 and the transition loops in Phase 2), 90 L of advanced feedwater was added to the reservoir and disinfected using free chlorine (Clorox bleach; 5% w/v) to target a CT of 45 mg-min L−1. Free chlorine was converted to chloramines via addition (in excess) and mixing of lab-grade ammonium chloride in the pipe loop reservoir, which simulated the use of chloramines in the full-scale distribution system. There were two short-term batch preparation protocols that deviated from standard operating procedures in Phase 1, including testing a seven-day recirculation period for all pipe loops and storing the conventional feedwater prior to use (more information in the ESI†). In total, there were 41 recirculation periods during the study.
2.4. Discrepancies in pipe loop operation conditions
Despite attempts to maintain consistent operating conditions across the five pipe loops and over time, there were three main discrepancies observed. These involved differences in temperature, conventional feedwater cell counts, and chlorine concentration. It is important to keep these differences in mind when interpreting the results. The first discrepancy observed was that the temperatures of the pipe loop bulk waters were lower in Phase 2 compared to Phase 1 (Fig. S3†), following a drop in the outside ambient air temperature from a seasonal change: fall to winter and early spring. The second discrepancy observed was that total and intact cell counts in the conventional feedwater declined during Phase 1 but slightly increased in the advanced feedwater throughout the study (Fig. S2†). The decrease in cell counts observed in the conventional feedwater could be from the decrease in temperature concurrent with the change in seasons. Other researchers have observed microbial water quality effects from changes in season in full-scale conventional drinking water distribution systems.31–33 The increase in cell counts observed in the advanced feedwater could be from longer residence times in the reservoir at the end of the study: the final three deliveries of RO permeate to the study site had an average of 14 days between deliveries compared to the overall average of 6.1 days between deliveries. The third discrepancy observed was that chlorine concentrations at the end of recirculation periods were frequently low (i.e., <0.2 mg L−1; Fig. S4A and Table S3†). Loops 3–5 did not have any total chlorine measurements at the end of recirculation periods that were below the method detection limit (0.02 mg L−1). In contrast, Loops 1 and 2 had total chlorine measurements at the end of recirculation periods that were more frequently below detection limit: 16.7% of Loop 1 samples in Phase 1, 9.1% of Loop 1 samples in Phase 2, 5.6% of Loop 2 samples in Phase 1, and 4.5% of Loop 2 samples in Phase 2 (Table S3†). The average total chlorine concentration at the end of recirculation periods was higher in Loop 3 during Phase 1 (0.56 mg L−1 as Cl2), Loop 4 during Phase 1 (0.50 mg L−1 as Cl2), and Loop 5 during Phase 2 (0.79 mg L−1 as Cl2) compared to other pipe loops and phases (range from 0.096 to 0.26 mg L−1 as Cl2; Table S3†).2.5. Field water quality analysis and grab sampling
Bulk water in the feedwaters and pipe loops were evaluated by various field and laboratory measurements of physicochemical variables (Table S1†). Temperature and pH (Electrode Sealed SJ F; Fisher Scientific) and residual chlorine (DPD method, HACH® pocket colorimeter II; Hach) were measured in daily grab samples from pipe loops. Other variables including major ions, heavy metals, and general characteristics were measured either semi-weekly or weekly from grab samples (Table S1†). The method detection limits for both the free and total chlorine were 0.02 mg L−1. A summary of the physical and chemical water quality parameters is presented in Table S4.†We sampled bulk water for cell counts by flow cytometry, ATP, batch growth assays, qPCR targets, and 16S rRNA gene amplicon sequencing analyses (Table S2†). Before sample collection, sample taps were disinfected with 70% ethanol, flushed (to remove cells sloughing from the tap fixture and piping) at maximum flow rate to achieve the following: at least five minutes of flush flow (full-scale drinking water distribution system, RO permeate, and conventional feedwater), at least two minutes of flush flow (advanced feedwater), and about 1 L of flush volume (pipe loop sampling ports). Grab samples of water for cell counts and ATP quantification were collected in 500 mL autoclave-sterilized glass bottles containing excess sodium thiosulfate to quench residual chlorine, transported on ice, and stored at 4 °C until further processing. Flow cytometry and ATP assays were completed within 48 hours of sample collection. For pipe loop bulk water samples, cell counts and ATP were quantified for 66% and 63% of recirculation periods respectively, and samples were collected at the end of a recirculation period (for 24 and 23 recirculation periods, respectively) or one day before the end of the recirculation period (for three recirculation periods) unless otherwise noted. The qPCR targets and 16S rRNA gene amplicon library preparation and sequencing were completed after ultrafiltration and further processing of bulk water biomass in the laboratory.
We performed batch growth assays on RO permeate samples collected after each step of conveyance, transportation, and storage before blending in pipe loop reservoirs as previously described with slight modifications.34 Briefly, growth potential was assessed by measuring the change in total cell counts in bulk water after incubation at 30 °C for five days. Prior to incubation, 1 mL of microbial inoculum (unfiltered bottled mineral water; Evian, France) was added to samples along with inorganic nutrient and mineral stock solutions to induce carbon-limiting conditions.35 Following incubation at 30 °C with mixing by inversion once daily, all samples were stored at 4 °C and processed within three days of collection. Carbon-free glassware, filters, consumables, and nutrient solutions were prepared as described previously.36
2.6. Dead-end ultrafiltration concentration of bulk water
For qPCR and 16S rRNA gene amplicon sequencing analyses, bulk water biomass was concentrated using dead-end ultrafiltration as previously described29,37 with different volumes concentrated depending on the biomass and volume available by sample type: pipe loop bulk water (50–120 L), pipe loop feedwaters (100–700 L), drinking water distribution system water (350–1000 L), and RO permeate (700–4000 L) as previously described.29 For pipe loop samples, bulk water biomass was only collected at the end of a recirculation period, and it was collected for 39% of the recirculation periods in the study. Briefly, ultrafilters (REXEED 25S, Henry Schein, Melville, NY) were soaked overnight in bovine calf serum (5% w/v) and rinsed via crossflow filtration with sample water prior to filtering the sample. After sample filtration, ultrafilters were transported to the laboratory on ice and backflushed with sterile backflush solution (0.5% w/v Tween 80, 0.01% w/v sodium polyphosphate, and 0.001% w/v Y-30 antifoam emulsion). Backflush was collected in an autoclaved 1 L glass bottle and was used for downstream secondary concentration using polyethylene glycol (PEG) flocculation. Field blanks for dead-end ultrafiltration field sampling consisted of ultrafilters that were processed alongside samples, including overnight soaking, backflushing, and secondary concentration. After overnight soaking, field blank ultrafilters were flushed with 1 L of autoclaved deionized water (via crossflow filtration) to remove the soaking solution, capped with sterilized caps, brought to the field, retained at ambient temperature during sample filtration, and then returned to the laboratory for parallel processing with field samples. Nine field blanks were included for 16S rRNA gene amplicon sequencing data analysis (see Section 2.12).2.7. Pipe wall biofilm sampling and analysis
Periodically, 0.3 m PVC segments (2.54 cm diameter) were removed from the pipe loops to harvest the biofilm, which was analyzed for ATP, qPCR targets, and 16S rRNA gene amplicon sequencing (Table S2†). Each biofilm sampling event occurred either at the end of a recirculation period (for six recirculation periods) or the day before the end of the recirculation period (for two recirculation periods), and biofilm was collected for 20% of recirculation periods in the study on the following dates: 12/13/2017, 12/29/2017, 1/5/2018, 1/11/2018, 1/15/2018, 1/18/2018, 2/15/2018, and 3/26/2018. 700 mL of water from the corresponding pipe loop was filtered (0.22 μm pore size syringe filter) and quenched with excess sodium thiosulfate. Isolation valves on either side of the segment were closed, and the entire segment exterior was wiped down with 70% ethanol to disinfect and remove accumulated dust/dirt. The segment was then isolated, removed by disconnecting the true-union fittings, drained, filled with the filtered water, capped on both ends, and transported on ice to the laboratory for further processing. The segment was replaced by a PVC segment that was disinfected (submerged in 0.5% bleach solution for >30 min) and rinsed with conventional feedwater. Each biofilm sampling event used a pipe segment that had been in place in the pipe loop since the beginning of operation: the replacement pipe segments were not sampled. Biofilm was recovered from the inside walls of the harvested pipe segments by a combination of two 3 min sonication periods (Branson 3510-DTH) and scraping. Full details are presented in the ESI.† In total, 160 to 260 mL of sample water was produced from the sonication procedure. A small aliquot (<10 mL) was analyzed for total and intracellular ATP concentrations, and the remaining sample underwent secondary concentration via PEG flocculation prior to DNA extraction and genomic analyses.2.8. PEG flocculation
PEG flocculation was used to concentrate sonicated water from biofilm samples, as well as ultrafilter backflush from bulk water samples and field blanks, following a protocol from Mark Borchardt at the United States Department of Agriculture (USDA; Marshfield, WI) as previously described with slight modifications.29,38 Briefly, backflush samples or sonicated biofilm samples were mixed with 1.15% w/v NaCl, 8% w/v polyethylene glycol 8000, and 1% w/v beef extract (catalog #DF0115173; Fisher Scientific). The solution was then kept at 4 °C as it was stirred for 1 hour, incubated overnight, transferred to autoclave-sterilized 500 mL centrifuge tubes, and centrifuged at 4200 RPM (relative centrifugal force not known) for 45 minutes in a swing-bucket centrifuge (Sorvall RC 5C with SH-3000 rotor). Supernatant was removed by decanting, and resulting pellets were resuspended in 1 to 4 mL of autoclave-sterilized tris-EDTA buffer, split into two equal aliquots (with volumes from 0.5 to 4.5 mL per aliquot), and stored at −80 °C until DNA extraction and genomic analyses.2.9. DNA extraction
DNA extraction from concentrated biomass samples was completed using a PowerSoil Pro extraction kit (Qiagen) with slight modifications, as previously described.29 Briefly, one pellet aliquot was thawed and vortexed for 10 seconds. For advanced feedwater, 200 μL of homogenized sample was added directly to each PowerSoil Pro Powerbead tube. For all other samples, sample homogenates were centrifuged at 34000 × g for 1 minute, and the pellet was stored on ice. To concentrate nucleic acids remaining in the supernatant, it was aliquoted onto a centrifugal filtration unit (Amicon ultra-15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 kDa; Millipore, Cork, Ireland). The filtration unit was centrifuged at 7500 × g for 30 minutes and the concentrate was combined with the pellet and homogenized. 200 μL of the concentrated sample (out of a total volume ranging from 0.25 to 2 mL) was added to the Powerbead Tube. The sample was incubated at 37 °C for 30 min with an enzymatic digestion solution: 50 μL of 0.001% lysozyme (Sigma-Aldrich, Darmstadt, Germany), 50 μL of 0.00001% achromopeptidase (Sigma-Aldrich, Darmstadt, Germany), and 8 μL of 0.01% carrier RNA in buffer AVL (Qiagen). Solution CD1 was added to the sample (500 μL), and then the PowerSoil Pro kit was followed as specified by the manufacturer until the step immediately before elution, when a 5 min room-temperature incubation step was added. The sample was eluted and stored at −80 °C until further processing. Six extraction blanks of nuclease-free water were included for 16S rRNA gene amplicon sequencing data analysis (see Section 2.12).
100 kDa; Millipore, Cork, Ireland). The filtration unit was centrifuged at 7500 × g for 30 minutes and the concentrate was combined with the pellet and homogenized. 200 μL of the concentrated sample (out of a total volume ranging from 0.25 to 2 mL) was added to the Powerbead Tube. The sample was incubated at 37 °C for 30 min with an enzymatic digestion solution: 50 μL of 0.001% lysozyme (Sigma-Aldrich, Darmstadt, Germany), 50 μL of 0.00001% achromopeptidase (Sigma-Aldrich, Darmstadt, Germany), and 8 μL of 0.01% carrier RNA in buffer AVL (Qiagen). Solution CD1 was added to the sample (500 μL), and then the PowerSoil Pro kit was followed as specified by the manufacturer until the step immediately before elution, when a 5 min room-temperature incubation step was added. The sample was eluted and stored at −80 °C until further processing. Six extraction blanks of nuclease-free water were included for 16S rRNA gene amplicon sequencing data analysis (see Section 2.12).
2.10. Flow cytometry, ATP analysis, and quantitative PCR
Total and intact cell counts were determined for bulk water grab samples as was previously described.11 Briefly, cell concentrations were measured using flow cytometry with SYBR Green I (S9430; Sigma-Aldrich, St. Louis, MO) and propidium iodide (30 mM P1304MP; Life Technologies, Carlsbad, CA) to distinguish cells with intact membranes. From each bulk water grab sample, three 1000 μL or 1500 μL aliquots were processed and the geometric mean and geometric standard deviation were calculated. Measurements were performed on two separate flow cytometers, an Accuri™ C6 flow cytometer (Accuri; BD Biosciences, San Jose, CA) and a BD FACSCanto™ cell analyzer (Canto; BD Biosciences, SanJose, CA), as previously described.30 The Accuri was used to sample until February 6th, 2018, and the Canto was used for the remaining study period. More detail can be found in the ESI.† For the Accuri, the limits of detection were previously determined for intact cell count (22 cells per mL) and total cell count (12 cells per mL) using the same instrument used in this study.11 The Canto limit of detection was based on a recommended lower quantification limit (102 cells per mL).39For ATP assays, the BacTiter-Glo™ kit (Promega Corporation) and a GloMax Luminometer (Model #E6080) were used as previously described.11 Briefly, total and intracellular ATP was measured in triplicate 500 μL aliquots of bulk water and biofilm samples. The geometric mean and geometric standard deviation were calculated. For total and extracellular ATP, the quantification limits were set by the standard curve (1 × 10−4 nM to 10 nM). For ATP assays as well as cell count assays, we were not able to complete all sample analyses for some locations on all sample days because of logistical constraints which resulted in small inconsistencies in the number of samples for each assay and location.
qPCR sequences for primers and probes were applied as described previously to target the 16S rRNA gene, Mycobacterium avium complex, Legionella pneumophila, Acanthamoeba spp., blaTEM, sul1 (Table S2†).29,40–43 Briefly, samples were analyzed in technical triplicate on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA) using triplicate DNA (gBlocks Gene Fragments; Integrated DNA Technologies, Coarlville, IA) standard curves on 96-well optical plates (MicroAmp™ Fast Optical). DNA standard curves on each qPCR plate were used to calculate gene counts and establish a limit of quantification (LoQ) for each assay. qPCR validation data (Tables S5, S6, and Fig. S5†) and standard details (Table S7†) can be found in the ESI.† Triplicate negative controls (i.e., PCR-grade water) were run on every plate. All negative controls amplified below the LoQ of each assay. Samples were considered positive (i.e., detected) if there was amplification in at least one of three qPCR triplicates, including if amplification was below the LoQ. For Legionella pneumophila, blaTEM, Mycobacterium avium complex, and Acanthamoeba spp., less than 22% of samples were within the quantifiable range, and only percent positive data are shown. For the 16S rRNA gene and sul1, the LoQs were determined to be 1000 and 10 gene copies per PCR reaction respectively. For statistical analyses and log10 reduction calculations, all values below the LoQ were set at the LoQ. Primers and probes for all assays were selected from the literature (Table S8†). There was insufficient extracted sample volume to complete all analyses on all samples, so subsets of samples representative of each phase were measured for each qPCR assay. Thermal cycling conditions for each assay were optimized before analysis of samples (Table S9†). Inhibition testing of samples followed the spike and dilute method44 (Table S10†). Detailed information on qPCR methodology is presented in the ESI.†
2.11. Library preparation and 16S rRNA gene amplicon sequencing
Library preparation for amplicon sequencing followed the Schloss Lab MiSeq wet-lab protocol for amplification of the V4 region of the 16S rRNA gene and Kantor et al. (2019),10 with slight modifications. Briefly, the V4 region was amplified using uniquely barcoded 515F and 806R primers with Phusion HotStart II polymerase (ThermoFisher Scientific), HF buffer (ThermoFisher Scientific), and 10 to 25 ng (up to 2 μL total) of genomic DNA. Triplicate 25 μL reactions were combined. For reactions that failed to amplify, PCR was repeated with 3% dimethyl sulfoxide and 0.2% bovine serum albumin added to the reaction mixture (25 μL total volume reactions). At the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, the dual-barcoded libraries were pooled and then sequenced on an Illumina MiSeq, yielding 300 bp paired-end reads. Two library preparation blanks of nuclease-free water (one on each plate) were included for 16S rRNA gene amplicon sequencing data analysis (see Data analysis section). The data have been deposited with links to BioProject accession number PRJNA896721 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).Amplicon sequence data were processed as previously described10 with slight modifications. Briefly, reads were demultiplexed, mapped to PhiX, and processed using DADA2 (v1.12.1)45 to generate amplicon sequence variants (ASVs). FastQC was used to assess quality and to determine cutoffs for the following five quality control measures: (i) truncation of all reads after 251 nucleotides (nts); (ii) trimming of all reads to remove 5 nts from the 5′ end; (iii) truncation of a subset of reads where quality score dropped to 10 or below; (iv) removal of reads with expected errors greater than 1; and (v) removal of reads with lengths less than 200 nts. The remaining reads were denoised, chimeras were removed from the dataset using removeBimeraDenovo, and taxonomy of sequences was assigned using the Ribosomal Database Project Naive Bayesian classifier46 trained using data from the SILVA database (v132).47
2.12. Data analysis
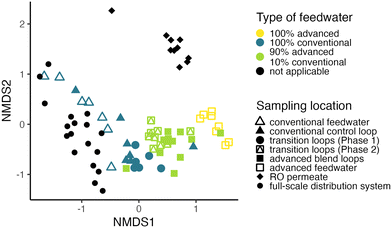
Data analysis was completed in R (v4.1.3) unless otherwise specified. Relative abundance of sul1 was calculated as the concentration of sul1 divided by the 16S rRNA gene concentration. Flow cytometric cell count data, intracellular ATP concentrations, and sul1 relative abundance data were not normally distributed; for comparisons between locations, a Kruskal–Wallis test was performed followed by pairwise Dunn's tests with Holm–Bonferroni correction using rstatix (v0.7).Data analysis for 16S rRNA gene amplicon sequencing was completed using Phyloseq (v1.38.0). Two blanks with fewer than 7500 reads were removed from downstream analyses. Contamination in samples was assessed using differential abundance analysis with DeSeq2 (v1.24.0) as previously described.10 In brief, samples were compared to extraction, library preparation, and field blanks and ASVs shared between samples and any negative control were determined. 151 ASVs were removed from the dataset that were shared between samples and any negative control and that were not significantly enriched in samples over negative controls. Subsequently, only samples with more than 300 reads remaining were kept for further analysis. The percent relative abundance of each ASV in each sample was determined and then Bray–Curtis dissimilarities were calculated. Non-metric multidimensional scaling (NMDS) of Bray–Curtis dissimilarities was performed for all bulk water samples as well as only pipe loop bulk water samples (i.e., not including samples from the full-scale distribution system, the demonstration-scale RO permeate, or the feedwater to the pipe loops). Clustering by sample date and feedwater composition was assessed in pipe loop bulk water samples using PERMANOVA (Vegan v2.5.7). Estimated absolute abundances were calculated for each sample by multiplying the relative abundance fraction with the total cell count measured. Differential abundance analysis was completed using DeSeq2 to identify significantly (p < 0.05) enriched or depleted ASVs after the pipe loops transitioned from conventional drinking water to the advanced blends. 16S rRNA gene amplicon sequencing data preparation code (https://github.com/rosekantor/awtp2) as well as 16S rRNA gene amplicon sequencing and water quality data analysis code (https://doi.org/10.5281/zenodo.7734207) are publicly available.
3. Results
3.1. Microbial water quality of feedwaters
The microbial water quality of the conventional feedwater samples was expected to closely resemble that of the full-scale drinking water distribution system samples because it originated from a distribution pipe onsite at the drinking water treatment facility and just downstream of treatment (Fig. 1A). Throughout the study period, a large portion of microbial cells in the conventional feedwater were intact (average total and intact cell counts of 6.79 × 104 and 1.98 × 104 cells per mL, respectively), despite a relatively high total chlorine concentration in the conventional feedwater (average of 2.20 mg L−1 as Cl2). Intracellular ATP (Fig. 2A), total cell counts (Fig. 2B), and intact cell counts (Fig. 2C) were not significantly different between the conventional feedwater and the full-scale drinking water distribution system samples further downstream.In contrast, the handling of advanced feedwater prior to use (Fig. 1A) resulted in a water quality that differed from the RO permeate at the advanced wastewater treatment plant and the conventional feedwater. While the RO permeate initially had very low cell counts, it was transported by tanker truck from the advanced wastewater treatment facility to the pipe loop site, conditioned to adjust the pH and alkalinity, and stored without disinfectant residual in a large outdoor plastic tank until batch chlorination (Fig. 1A). The average water age of the conditioned, stored RO permeate (advanced feedwater) was 6.1 ± 5.4 days, which provided substantial time for microorganisms to grow prior to batch chlorination. Intracellular ATP (Fig. 2A), total cell counts (Fig. 2B), and intact cell counts (Fig. 2C) were significantly higher in the advanced feedwater (prior to batch chlorination) compared to the RO permeate samples. In addition, the microbial community shifted during handling: RO permeate samples clustered together tightly and separately from advanced feedwater (Fig. 3). Microbial batch growth assays conducted under carbon-limiting conditions were completed during each step of RO permeate handling to determine if bioavailable carbon was introduced during RO handling. Each handling step, including conveyance, transportation (highest contribution), and storage onsite at the conventional drinking water treatment facility, introduced bioavailable carbon (Table S11†). As a result of the RO permeate handling, there was a shift in the microbial community composition (Fig. 3) and a >1000-fold increase in the average concentration of intact cells to levels that were significantly higher than the conventional feedwater (Fig. 2).
During the final step to produce the advanced blend for the pipe loops, batch chlorination was used to disinfect 90 L of advanced feedwater and to yield a chloramine residual in the pipe loop reservoirs prior to blending with 10 L of conventional feedwater. Although we did not sample this blend prior to its first introduction to the pipe loops, the samples from the advanced blend pipe loops (after a 3.5 day recirculation period) provide insight into its water quality. As discussed further in the next section, the batch chlorination of the RO permeate that had been transported, conditioned and stored led to a feedwater quality that was unlike typical finished drinking water due to the high concentration of bacterial cells damaged by chlorination.
3.2. Microbial water quality in the conventional and advanced blend pipe loops
Three pipe loops were fed with a consistent feedwater composition throughout the study: the conventional control loop and two advanced blend loops (Fig. 1B). This section focuses on a comparison of these three loops, prior to presenting the results for the loops that were transitioned from 100% conventional feedwater to advanced blends (Section 3.3). The conventional control loop was intended to represent the full-scale drinking water distribution system. Intracellular ATP (Fig. 2A), total cell counts (Fig. 2B), and intact cell counts (Fig. 2C) were not significantly different between the conventional control loop, the conventional feedwater, and the full-scale drinking water distribution system samples. These results indicate that the conventional feedwater was fairly “stable” with respect to microbial growth, because cell counts did not increase in the pipe loops (after 3.5 days of recirculation) nor were cell counts higher in the full-scale distribution system samples. In addition, the microbial community of the conventional control loop was more similar to that of the full-scale distribution system compared to the RO permeate (Fig. 3).The advanced blend loops had significantly lower concentrations of intracellular ATP, total cell counts, and intact cell counts than the (unchlorinated) advanced feedwater. This finding suggests that disinfection caused both loss of membrane integrity and lysis of cells (Fig. 2). We expected an inverse relationship between chlorine residual and intact cell counts, as has been observed in chlorinated and chloraminated full-scale drinking water distribution systems.11,30,34,48–50 Instead, we observed a decline in both chlorine concentration and intact cell counts (Fig. S6†). We hypothesized that, in the advanced blend loops, cells continued to lyse throughout each 3.5 day recirculation period rather than grow in response to decreased disinfectant concentrations. This trend was not reflective of what has been observed previously in the same full-scale conventional drinking water distribution system,30 and provides insight into the unique conditions that were created by disinfecting the advanced feedwater with high cell counts from recent growth. Nonetheless, the microbial community of the advanced blend loops resembled that of the advanced feedwater (Fig. 3), but it is possible that we would have observed a shift in the microbial community if DNA from only intact cells was sequenced.
Interestingly, the advanced blend loops had similar intracellular ATP and intact cell counts to that of the full-scale distribution system, conventional feedwater, and conventional control loop (Fig. 2A and C). However, the total cell count was significantly higher (Fig. 2B), which also suggests cell lysis continued to occur as the advanced blend feed recirculated in the pipe loops. While the conventional and advanced feedwaters were characterized by distinct microbial community profiles, the microbial community profiles in the advanced blend loops converged between the feedwaters (Fig. 3). Of note, average total chlorine concentrations in the advanced blend loops at the end of recirculation periods were frequently low in Phase 2 (≥50% of samples had <0.2 mg L−1 as Cl2; Table S3†) at the end of recirculation periods as compared to the conventional control loop in Phase 2 (average of 0.79 mg L−1 as Cl2; Fig. S4 and Table S3†). Disinfectant residual has been shown to affect microbial water quality in drinking water distribution systems,30,51–57 and there is an inherent challenge of comparing test conditions with different disinfectant residual concentrations.
3.3. Pipe loop transition from conventional feedwater to the advanced blend
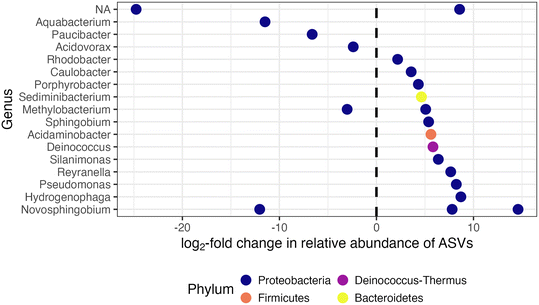
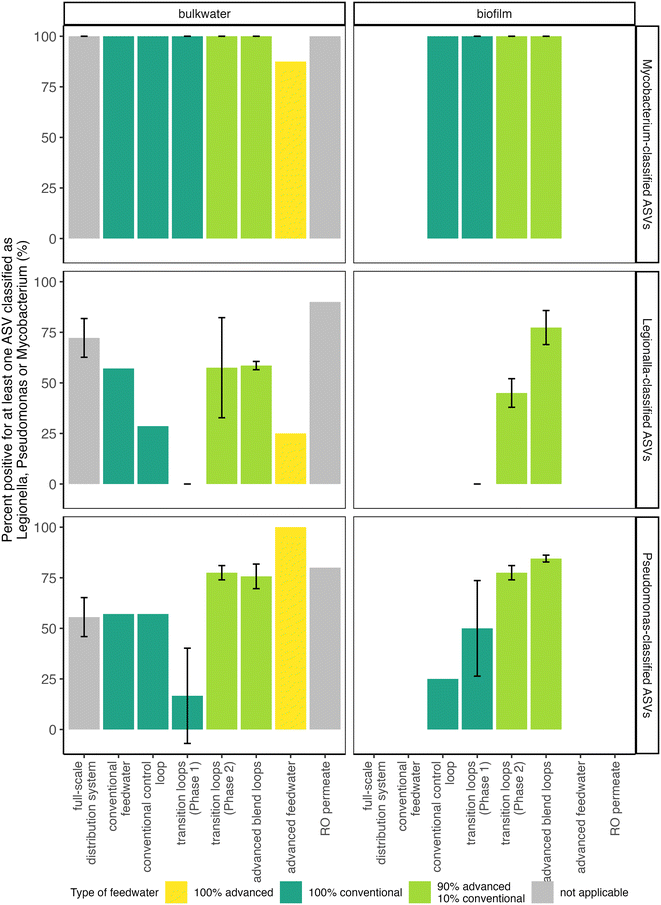
Two pipe loops were transitioned from conventional feedwater (Phase 1) to advanced blend feedwater (Phase 2; Fig. 1B). To assess biofilm sloughing in the transition loops, we determined if there was a significant increase in intracellular ATP or intact cell counts in bulk water or decrease in intracellular ATP in Phase 2 compared to Phase 1. We did not observe evidence of biofilm sloughing: intact and total cell counts in the bulk water of the transition loops were not significantly different in Phase 1 compared to Phase 2 (Fig. S4B†), nor was the intracellular ATP of biofilm samples (Fig. 2A). Indeed, the intracellular ATP in bulk water and biofilm, and the intact cell counts in the transition loop bulk water were similar to that of the conventional control loop (Fig. 2A and C). The timescale for chemical and biological destabilization in drinking water distribution systems is not well established,17 but we did not observe large increases in intact cell count in the pipe loop bulk water that were immediate (after the first 3.5 day recirculation period) or long-term (during the 11 weeks of Phase 2; Fig. S4b†).The feedwater type affected the microbial community profiles in the bulk water and biofilm of the pipe loops. First, the effect of feedwater type on the microbial community profiles of all pipe loop bulk water samples collected in this study was assessed (Fig. S7A†). In the transition loops, the microbial community profile shifted in Phase 2 and samples clustered between the conventional and advanced blend loops. The pipe loop bulk water samples clustered by type of feedwater (R2 = 0.24, p = 0.001), but sample date did not explain the variation in microbial community profiles (R2 = 0.03, p < 0.05). Next, the effect of sample type (i.e., bulk water compared to biofilm) compared to feedwater type on the microbial community profiles in the transition loops was assessed (Fig. S7B†). The transition loop samples clustered visually by feedwater type, but feedwater type (R2 = 0.05, p < 0.05), sample type (R2 = 0.09, p < 0.01), and sample date (R2 = 0.04, p > 0.05) did not explain the variation in microbial community profiles. In the transition loops, 21 ASVs were significantly enriched or depleted after the feedwater switch to advanced blends from 100% conventional feedwater (Phase 2 compared to Phase 1; Fig. 4). Many of the enriched ASVs were introduced by the advanced feedwater (Fig. S8†). For example, the Hydrogenophaga-classified ASV was enriched in the transition loops in Phase 2 (Fig. 4) and had high relative abundance in most of the advanced feedwater samples (Fig. S8A†). Several of the genera enriched in Phase 2 have been identified in drinking water treatment and distribution systems in previous studies.15,58–61 The enriched Pseudomonas-classified ASV is discussed in more detail in the next sections.
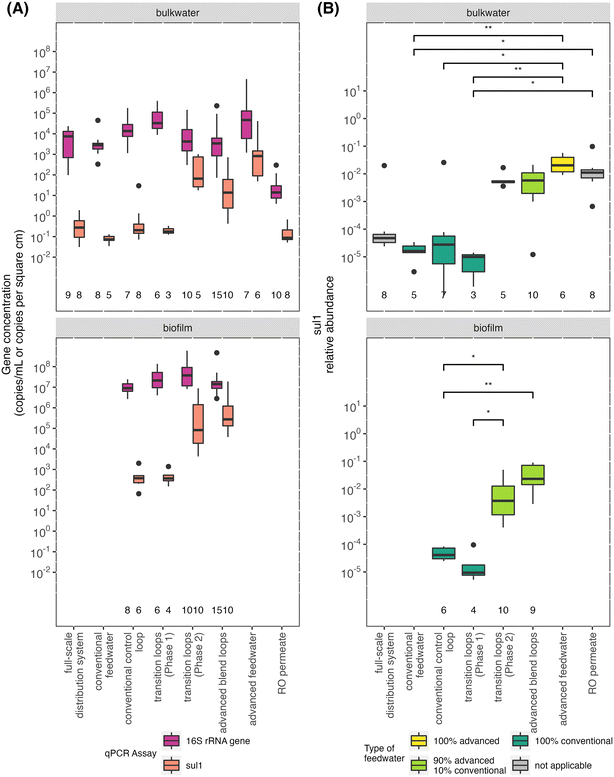
3.4. Antibiotic resistance genes sul1 and blaTEM
Sul1 concentrations were similar in the conventional feedwater, RO permeate, and full-scale distribution system and comparable to other previously studied full-scale distribution systems (Fig. 5A).9,62,63 In comparison to the conventional feedwater, the relative abundance (i.e., sul1 concentration/16S rRNA gene concentration) of sul1 in RO permeate was significantly higher (Fig. 5B). The RO permeate handling resulted in increased concentrations of the 16S rRNA gene and sul1. Sul1 increased proportionally such that the relative abundance remained consistently high in the advanced feedwater.The feedwater type affected the sul1 abundance measured in the bulk water and biofilm of the pipe loops. The pipe loops with the same feedwater type in Phases 1 and 2 had similar low (conventional control loop) or high (advanced blend loops) concentration and relative abundance of sul1 as the respective primary feedwater. Similarly, in the transition loops, the abundance of sul1 reflected that of the primary feedwater for each phase; importantly, the concentration and relative abundance of sul1 increased in both the bulk water and biofilm after introduction of the advanced blend (i.e., in Phase 2 compared to Phase 1; Fig. 5). Previous studies of drinking water have found that chlorine exposure reduced the relative abundance of sul1, but increased the relative abundance of other antibiotic resistance genes.62,64,65 We did not observe a significant difference in sul1 concentrations in the advanced feedwater compared to the pipe loops fed with the advanced blend (i.e., after the advanced feedwater underwent primary disinfection with chlorine, formation of chloramine residual, blending with conventional water, and recirculation for 3.5 days). However, the study design did not allow for measurement of sul1 concentrations immediately following disinfection because batch chlorination was completed in pipe loop reservoirs with water needed for subsequent bulk water filtration sampling events (90 L).
We observed a Pseudomonas-classified ASV that was enriched in the transition loops after the introduction of the advanced blend (Fig. 4), and Pseudomonas have been shown to carry sul1 (there are currently seven species of Pseudomonas with resistomes that include sul1 in the CARD database).66 The relative abundance of the enriched Pseudomonas-classified ASV was low in RO permeate samples (Fig. S9†) and higher in biofilm samples from pipe loops fed with the advanced blend (Fig. S8B†). With this dataset, it is not possible to determine if the Pseudomonas-classified ASV carried sul1, but Pseudomonas species in direct potable reuse systems and their potential for antibiotic resistance warrant further study.
The beta-lactam resistance gene blaTEM was rarely detected in this study. While sul1 was detected in all samples (n = 84), blaTEM was detected in only 33% of samples (28 of 84 samples). Of these, 15 samples were in the quantifiable range. All sample locations had at least one blaTEM detection, except the RO permeate samples (0 detects out of 7 tested).
3.5. Opportunistic pathogens assessed with 16S rRNA gene amplicon sequencing and qPCR
Mycobacterium spp. were present in nearly every sample collected in this study regardless of feedwater type, and Mycobacterium avium complex was detected via qPCR in the majority of samples tested. At least one Mycobacterium-classified ASV was present in all samples, except for some of the advanced feedwater samples (Fig. 6 and S10†), but no Mycobacterium-classified ASV was significantly enriched in the bulk water and biofilm of the transition loops in Phase 2 compared to Phase 1 (Fig. 4). In addition, 59% of samples tested were positive for the Mycobacterium avium complex gene by qPCR (40 of 68 samples), with only 2 samples in the quantifiable range. RO permeate samples were not tested for Mycobacterium avium complex, but all other sample locations had at least one sample positive for Mycobacterium avium complex by qPCR. The relatively frequent detection of Mycobacterium avium complex and Mycobacterium-classified ASVs in this study is consistent with previous studies in which Mycobacterium spp. has been observed in chloraminated full-scale distribution systems43,67 and in simulated distribution systems fed with advanced-treated wastewater.8,10Legionella spp. and Acanthamoeba spp. were present in many of the samples in this study, but Legionella pneumophila was rarely detected by qPCR. At least one Legionella-classified ASV was observed in a majority of samples and locations (Fig. 6), including the RO permeate. However, the majority of RO permeate samples only had one Legionella-classified ASV, and the estimated absolute abundance (i.e., the relative abundance multiplied by the intact cell count) of this ASV was relatively low in the majority of samples of the bulk water of the advanced blend loops (Legionella 144; Fig. S10†). Similarly, Acanthamoeba spp., a host of Legionella,21 was detected by qPCR in 46% of samples (36 out of 79 samples), including at least one detection for each sample location. In addition, biofilms of pipe loops fed with the advanced blend had more samples with at least one Legionella-classified ASV compared to biofilms from pipe loops fed with conventional feedwater (Fig. 6), but no Legionella-classified ASV was significantly enriched in the bulk water and biofilm of the transition loops in Phase 2 compared to Phase 1 (Fig. 4). Despite the presence of Legionella-classified ASVs and the detection of Acanthamoeba spp. via qPCR, Legionella pneumophila was detected by qPCR in only one advanced blend loop sample out of 73 samples tested (RO permeate samples were not tested by qPCR for Legionella pneumophila or Acanthamoeba spp.). These results are consistent with previous studies, which have documented low detection of Legionella pneumophila in chloraminated drinking water distribution systems.68,69
Pseudomonas spp. were present in the RO permeate and increased in pipe loop samples, but the proportion of Pseudomonas aeruginosa specifically is unknown. At least one Pseudomonas-classified ASV was present in the majority of samples collected from RO permeate, the advanced feedwater, and pipe loops fed with the advanced blend (Fig. 6). Similarly, more samples with at least one Pseudomonas-classified ASV were present in biofilms of pipe loops fed with the advanced blend compared to conventional feedwater (Fig. 6). In the advanced feedwater, two Pseudomonas-classified ASVs increased in estimated absolute abundance from the RO permeate, and one ASV was also in high abundance in the advanced blend pipe loops (“Pseudomonas_24”; Fig. S10†). This Pseudomonas-classified ASV was previously identified to be enriched in the transition loops after the introduction of the advanced blend (Fig. 4). We did not quantify Pseudomonas aeruginosa via qPCR in this study, but Pseudomonas aeruginosa was not detected by Garner et al. (2019)8 in simulated distribution systems fed with advanced-treated wastewater. Indeed, they found that simulated distribution systems fed with advanced-treated wastewater did not support the growth of any opportunistic pathogens quantified.
4. Discussion
4.1. Pipe loop transition from conventional feedwater to the advanced blend
In the transition loops, the transition from conventional feedwater to the advanced blend affected sul1 concentrations and the microbial community composition, but not absolute abundance metrics (e.g., ATP or cell counts). The microbial community composition of the transition loops reflected the respective primary feedwater before and after the transition, and advanced blend loops and conventional control loops reflected the respective primary feedwater throughout the study. A literature review did not reveal work that simulated the microbial impacts of the transition from conventional to treated water augmentation in distribution systems. However, other researchers have observed similar shifts in microbial community profiles in conventional systems following the transition to a new feedwater or upgraded treatment at full scale.14,15,23 21 ASVs were significantly enriched or depleted in the bulk water and biofilm following the transition, including one Pseudomonas-classified ASV that was also at high estimated absolute abundance in pipe loops fed with the advanced blend and not identified in the pipe loops fed with 100% conventional water. This Pseudomonas-classified ASV increased in estimated absolute abundance after handling of RO permeate. Similarly, sul1 in the bulk water and biofilm of the transition loops increased in both absolute and relative abundance after the transition, reflecting the composition of sul1 in the feedwaters. These findings indicate that bacteria from the feedwater were able to compete with the biofilm microbial community that was established prior to the transition. Chen et al. (2020)70 found higher relative abundance of sul1 in bulk water and biofilms in a conventional drinking water distribution system compared to the finished water at the drinking water treatment facility. In this study, we observed a large increase in the relative abundance of sul1 and the estimated absolute abundance of a Pseudomonas-classified ASV during handling of RO permeate prior to introduction in the pipe loops, and these findings support that bacterial growth during storage and conveyance of advanced-treated wastewater could increase antibiotic resistance and opportunistic pathogen estimated absolute abundance.4.2. Considerations for full-scale direct potable reuse systems
4.3. Considerations for future direct potable reuse pilot- and bench-scale studies
5. Conclusions
• The microbial community profile in the pipe loop bulk water and biofilm reflected that of the current primary feedwater, including in the pipe loops that transitioned from conventional water to direct potable reuse water.• The bioavailable carbon introduced during conveyance, transportation, and storage of RO permeate contributed to an increase in absolute microbial abundance, which included increases in antibiotic resistance and opportunistic pathogen estimated absolute abundance.
• Enhanced monitoring is recommended for full-scale systems that includes absolute microbial abundance, antibiotic resistance gene targets, and opportunistic pathogen targets for at least two years before and two years after transition to direct potable reuse.
• Future bench- or pilot-scale studies of direct potable reuse microbiology should be designed to include bulk water volume and biofilm sampling area that yield sufficient microbial biomass for analyses, appropriate simulated drinking water distribution systems for the study objectives, and comprehensive technical and experimental controls.
Publication disclaimer
This publication was developed under STAR Fellowship Assistance Agreement no. 91782901-0, awarded by the EPA. It has not been formally reviewed by EPA. The views expressed in this publication are solely those of the listed authors, and EPA does not endorse any products or commercial services mentioned in this publication.Author contributions
Lauren C. Kennedy: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, validation, visualization, writing – original draft, writing – review & editing. Scott E. Miller: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, validation, visualization, writing – original draft, writing – review & editing. Rose S. Kantor: conceptualization, data curation, funding acquisition, investigation, methodology, validation, writing – original draft, writing – review & editing. Hannah Greenwald: data curation, investigation, methodology, validation, writing – review & editing. Michael J. Adelman: conceptualization, data curation, funding acquisition, investigation, methodology, resources, writing – review & editing. Hari Seshan: conceptualization, data curation, funding acquisition, investigation, methodology, resources, writing – review & editing. Paige Russell: conceptualization, data curation, funding acquisition, investigation, methodology, resources, writing – review & editing. Kara L. Nelson: conceptualization, funding acquisition, methodology, resources, supervision, writing – original draft, writing – review & editing.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Science Foundation (NSF) Engineering Research Center for Re-Inventing the Nation's Urban Water Infrastructure [EEC-1028968] and a donation from our utility partner. L. C. K was supported by the NSF Graduate Research Fellowship [DGE-1752814]. S. E. M. was supported by the U.S. Environmental Protection Agency (EPA) STAR Fellowship Assistance Agreement no. 91782901-0.References
- G. P. Maseeh, C. G. Russell, S. L. Villalobos, J. E. Balliew and G. Trejo, El Paso's Advanced Water Purification Facility: A New Direction in Potable Reuse, J. AWWA, 2015, 107, 36–45 CrossRef.
- B. M. Pecson, S. C. Triolo, S. Olivieri, E. C. Chen, A. N. Pisarenko, C.-C. Yang, A. Olivieri, C. N. Haas, R. S. Trussell and R. R. Trussell, Reliability of pathogen control in direct potable reuse: Performance evaluation and QMRA of a full-scale 1 MGD advanced treatment train, Water Res., 2017, 122, 258–268 CrossRef CAS PubMed.
- J. A. Soller, S. E. Eftim, I. Warren and S. P. Nappier, Evaluation of microbiological risks associated with direct potable reuse, Microb. Risk Anal., 2016, 5, 3–14 CrossRef.
- E. Steinle-Darling, J. Sutherland and A. Salveson, Sampled Direct Potable Reuse Water Shows Promising Results, Opflow, 2016, 42, 20–22 CrossRef.
- E. L. Marron, W. A. Mitch, U. von Gunten and D. L. Sedlak, A Tale of Two Treatments: The Multiple Barrier Approach to Removing Chemical Contaminants During Potable Water Reuse, Acc. Chem. Res., 2019, 52, 615–622 CrossRef CAS PubMed.
- A proposed framework for regulating direct potable reuse in California, California Water Boards, 2018 Search PubMed.
- M. J. Adelman, J. H. Borchardt, H. Seshan, P. J. Russell, V. Occiano and A. M. Zare, Understanding system-level operational impacts in potable reuse planning: case studies from San Diego, CA, USA., 2021 Search PubMed.
- E. Garner, M. Inyang, E. Garvey, J. Parks, C. Glover, A. Grimaldi, E. Dickenson, J. Sutherland, A. Salveson, M. A. Edwards and A. Pruden, Impact of blending for direct potable reuse on premise plumbing microbial ecology and regrowth of opportunistic pathogens and antibiotic resistant bacteria, Water Res., 2019, 151, 75–86 CrossRef CAS PubMed.
- A. Olivieri, J. Crook, M. Anderson, R. Bull, J. Drewes, W. Jakubowski, P. McCarty, K. Nelson, J. Rose, D. Sedlak and T. Wade, Expert Panel Final Report: Evaluation of the Feasibility of Developing Uniform Water Recycling Criteria for Direct Potable Reuse, National Water Research Institute, 2016 Search PubMed.
- R. S. Kantor, S. E. Miller and K. L. Nelson, The Water Microbiome Through a Pilot Scale Advanced Treatment Facility for Direct Potable Reuse, Front. Microbiol., 2019, 10, 21 CrossRef PubMed.
- S. E. Miller, R. A. Rodriguez and K. L. Nelson, Removal and growth of microorganisms across treatment and simulated distribution at a pilot-scale direct potable reuse facility, Environ. Sci.: Water Res. Technol., 2020, 107, 36 Search PubMed.
- M. J. Adelman, M. S. Smith, T. D. Hancock, J. H. Borchardt, M. D. Williams, J. Quicho and M. L. Weber-Shirk, A bench-scale study of potable reuse impacts on surface water treatment, AWWA Water Sci., 2020, 2, e1205 CAS.
- M. C. Meckes, R. C. Haught, K. Kelty, J. C. Blannon and D. Cmehil, Impact on water distribution system biofilm densities from reverse osmosis membrane treatment of supply water, J. Environ. Eng. Sci., 2007, 6, 449–454 CrossRef CAS.
- L. Chen, F. Ling, G. Bakker, W.-T. Liu, G. Medema, W. van der Meer and G. Liu, Assessing the transition effects in a drinking water distribution system caused by changing supply water quality: an indirect approach by characterizing suspended solids, Water Res., 2020, 168, 115159 CrossRef CAS PubMed.
- Y. Hu, D. Dong, K. Wan, C. Chen, X. Yu and H. Lin, Potential shift of bacterial community structure and corrosion-related bacteria in drinking water distribution pipeline driven by water source switching, Front. Environ. Sci. Eng., 2020, 15, 28 CrossRef.
- I. Sibille, L. Mathieu, J. L. Paquin, D. Gatel and J. C. Block, Microbial characteristics of a distribution system fed with nanofiltered drinking water, Water Res., 1997, 31, 2318–2326 CrossRef CAS.
- G. Liu, Y. Zhang, W.-J. Knibbe, C. Feng, W. Liu, G. Medema and W. van der Meer, Potential impacts of changing supply-water quality on drinking water distribution: A review, Water Res., 2017, 116, 135–148 CrossRef CAS PubMed.
- P. Laurent, P. Servais, D. Gatel, G. Randon, P. Bonne and J. Cavard, Microbiological quality before and after nanofiltration, J. AWWA, 1999, 91, 62–72 CrossRef CAS.
- E. Torvinen, S. Suomalainen, M. J. Lehtola, I. T. Miettinen, O. Zacheus, L. Paulin, M.-L. Katila and P. J. Martikainen, Mycobacteria in Water and Loose Deposits of Drinking Water Distribution Systems in Finland, Appl. Environ. Microbiol., 2004, 70, 1973 CrossRef CAS PubMed.
- D. van der Kooij, G. L. Bakker, R. Italiaander, H. R. Veenendaal and B. A. Wullings, Biofilm Composition and Threshold Concentration for Growth of Legionella pneumophila on Surfaces Exposed to Flowing Warm Tap Water without Disinfectant, Appl. Environ. Microbiol., 2017, 83 DOI:10.1128/AEM.02737-16.
- N. J. Ashbolt, Environmental (Saprozoic) Pathogens of Engineered Water Systems: Understanding Their Ecology for Risk Assessment and Management, Pathogens, 2015, 4, 390–405 CrossRef PubMed.
- Water Services Association of Australia, All options on the table: Lessons learned from the journeys of others, 2019 Search PubMed.
- S. Chan, K. Pullerits, A. Keucken, K. M. Persson, C. J. Paul and P. Radström, Bacterial release from pipe biofilm in a full-scale drinking water distribution system, npj Biofilms Microbiomes, 2019, 5, 9–9 CrossRef PubMed.
- I. B. Gomes, M. Simões and L. C. Simões, An overview on the reactors to study drinking water biofilms, Water Res., 2014, 62, 63–87 CrossRef CAS PubMed.
- A. C. Martiny, T. M. Jørgensen, H.-J. Albrechtsen, E. Arvin and S. Molin, Long-Term Succession of Structure and Diversity of a Biofilm Formed in a Model Drinking Water Distribution System, Appl. Environ. Microbiol., 2003, 69, 6899–6907 CrossRef CAS PubMed.
- I. Douterelo, K. E. Fish and J. B. Boxall, Succession of bacterial and fungal communities within biofilms of a chlorinated drinking water distribution system, Water Res., 2018, 141, 74–85 CrossRef CAS PubMed.
- I. Douterelo, R. Sharpe and J. Boxall, Bacterial community dynamics during the early stages of biofilm formation in a chlorinated experimental drinking water distribution system: implications for drinking water discolouration, J. Appl. Microbiol., 2014, 117, 286–301 CrossRef CAS PubMed.
- K. Garny, H. Horn and T. R. Neu, Interaction between biofilm development, structure and detachment in rotating annular reactors, Bioprocess Biosyst. Eng., 2008, 31, 619–629 CrossRef CAS PubMed.
- S. Miller, H. Greenwald, L. C. Kennedy, R. S. Kantor, R. Jiang, A. Pisarenko, E. Chen and K. L. Nelson, Microbial Water Quality through a Full-Scale Advanced Wastewater Treatment Demonstration Facility, ACS ES&T Engg, 2022, 2(12), 2206–2219 Search PubMed.
- L. C. Kennedy, S. E. Miller, R. S. Kantor and K. Nelson, Effect of disinfectant residual, pH, and temperature on microbial abundance in disinfected drinking water distribution systems, Environ. Sci.: Water Res. Technol., 2021, 7(78), 78–92 RSC.
- A. J. Pinto, J. Schroeder, M. Lunn, W. Sloan and L. Raskin, Spatial-Temporal Survey and Occupancy-Abundance Modeling To Predict Bacterial Community Dynamics in the Drinking Water Microbiome, MBio, 2014, 5 DOI:10.1128/mBio.01135-14.
- F. Ling, C. Hwang, M. W. LeChevallier, G. L. Andersen and W.-T. Liu, Core-satellite populations and seasonality of water meter biofilms in a metropolitan drinking water distribution system, ISME J., 2015, 10, 582–595 CrossRef PubMed.
- E. I. Prest, D. G. Weissbrodt, F. Hammes, M. C. M. van Loosdrecht and J. S. Vrouwenvelder, Long-Term Bacterial Dynamics in a Full-Scale Drinking Water Distribution System, PLoS One, 2016, 11, e0164445 CrossRef CAS PubMed.
- S. Gillespie, P. Lipphaus, J. Green, S. Parsons, P. Weir, K. Juskowiak, B. Jefferson, P. Jarvis and A. Nocker, Assessing microbiological water quality in drinking water distribution systems with disinfectant residual using flow cytometry, Water Res., 2014, 65, 224–234 CrossRef CAS PubMed.
- E. I. Prest, F. Hammes, M. C. M. van Loosdrecht and J. S. Vrouwenvelder, Biological Stability of Drinking Water: Controlling Factors, Methods, and Challenges, Front. Microbiol., 2016, 7, 133 Search PubMed.
- F. A. Hammes and T. Egli, New Method for Assimilable Organic Carbon Determination Using Flow-Cytometric Enumeration and a Natural Microbial Consortium as Inoculum, Environ. Sci. Technol., 2005, 39, 3289–3294 CrossRef CAS PubMed.
- C. M. Smith and V. R. Hill, Dead-End Hollow-Fiber Ultrafiltration for Recovery of Diverse Microbes from Water, Appl. Environ. Microbiol., 2009, 75, 5284–5289 CrossRef PubMed.
- M. A. Borchardt, N. L. Haas and R. J. Hunt, Vulnerability of Drinking-Water Wells in La Crosse, Wisconsin, to Enteric-Virus Contamination from Surface Water Contributions, Appl. Environ. Microbiol., 2004, 70, 5937–5946 CrossRef CAS PubMed.
- E. Gatza, F. Hammes and E. Prest, Assessing water quality with the BD Accuri C6 flow cytometer, BD Biosciences, 2013, 1–12 Search PubMed.
- P. H. Dobrowsky, S. Khan and W. Khan, Resistance of Legionella and Acanthamoeba mauritaniensis to heat treatment as determined by relative and quantitative polymerase chain reactions, Environ. Res., 2017, 158, 82–93 CrossRef CAS PubMed.
- L. Proia, A. Anzil, J. Subirats, C. Borrego, M. Farrè, M. Llorca, J. L. Balcázar and P. Servais, Antibiotic resistance along an urban river impacted by treated wastewaters, Sci. Total Environ., 2018, 628–629, 453–466 CrossRef CAS PubMed.
- S. S. Silkie and K. L. Nelson, Concentrations of host-specific and generic fecal markers measured by quantitative PCR in raw sewage and fresh animal feces, Water Res., 2009, 43, 4860–4871 CrossRef CAS PubMed.
- H. Whiley, A. Keegan, H. Fallowfield and R. Bentham, Detection of Legionella, L. pneumophila and Mycobacterium Avium Complex (MAC) along Potable Water Distribution Pipelines, Int. J. Environ. Res. Public Health, 2014, 11, 7393–7405 CrossRef PubMed.
- Y. Cao, J. F. Griffith, S. Dorevitch and S. B. Weisberg, Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters, J. Appl. Microbiol., 2012, 113, 66–75 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- Q. Wang, G. M. Garrity, J. M. Tiedje and J. R. Cole, Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy, Appl. Environ. Microbiol., 2007, 73, 5261–5267 CrossRef CAS PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glöckner, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2013, 41, D590–D596 CrossRef CAS PubMed.
- C. Bertelli, S. Courtois, M. Rosikiewicz, P. Piriou, S. Aeby, S. Robert, J.-F. Loret and G. Greub, Reduced Chlorine in Drinking Water Distribution Systems Impacts Bacterial Biodiversity in Biofilms, Front. Microbiol., 2018, 9, 2496 CrossRef PubMed.
- R. Cheswick, E. Cartmell, S. Lee, A. Upton, P. Weir, G. Moore, A. Nocker, B. Jefferson and P. Jarvis, Comparing flow cytometry with culture-based methods for microbial monitoring and as a diagnostic tool for assessing drinking water treatment processes, Environ. Int., 2019, 130, 104893 CrossRef PubMed.
- A. Nescerecka, J. Rubulis, M. Vital, T. Juhna and F. Hammes, Biological Instability in a Chlorinated Drinking Water Distribution Network, PLoS One, 2014, 9, e96354 CrossRef PubMed.
- Q. M. Bautista-de los Santos, J. L. Schroeder, M. C. Sevillano-Rivera, R. Sungthong, U. Z. Ijaz, W. T. Sloan and A. J. Pinto, Emerging investigators series: microbial communities in full-scale drinking water distribution systems – a meta-analysis, Environ. Sci.: Water Res. Technol., 2016, 2, 631–644 RSC.
- J. Chen, W. Li, Q. Tan, D. Sheng, Y. Li, S. Chen and W. Zhou, Effect of disinfectant exposure and starvation treatment on the detachment of simulated drinking water biofilms, Sci. Total Environ., 2022, 807, 150896 CrossRef CAS PubMed.
- Z. Dai, M. C. Sevillano-Rivera, S. T. Calus, Q. M. Bautista-de los Santos, A. M. Eren, P. W. J. J. van der Wielen, U. Z. Ijaz and A. J. Pinto, Disinfection exhibits systematic impacts on the drinking water microbiome, Microbiome, 2020, 8, 42 CrossRef CAS PubMed.
- B. Hegarty, Z. Dai, L. Raskin, A. Pinto, K. Wigginton and M. Duhaime, A snapshot of the global drinking water virome: Diversity and metabolic potential vary with residual disinfectant use, Water Res., 2022, 218, 118484 CrossRef CAS PubMed.
- J. Inkinen, S. Siponen, B. Jayaprakash, A. Tiwari, A.-M. Hokajärvi, A. Pursiainen, J. Ikonen, A. Kauppinen, I. T. Miettinen, J. Paananen, E. Torvinen, M. Kolehmainen and T. Pitkänen, Diverse and active archaea communities occur in non-disinfected drinking water systems–Less activity revealed in disinfected and hot water systems, Water Res.: X, 2021, 12, 100101 CAS.
- A. Nescerecka, T. Juhna and F. Hammes, Identifying the underlying causes of biological instability in a full-scale drinking water supply system, Water Res., 2018, 135, 11–21 CrossRef CAS PubMed.
- C. Thom, C. J. Smith, G. Moore, P. Weir and U. Z. Ijaz, Microbiomes in drinking water treatment and distribution: A meta-analysis from source to tap, Water Res., 2022, 212, 118106 CrossRef CAS PubMed.
- H. Wang, C. Hu and B. Shi, The control of red water occurrence and opportunistic pathogens risks in drinking water distribution systems: A review, J. Environ. Sci., 2021, 110, 92–98 CrossRef CAS PubMed.
- I. Vaz-Moreira, O. C. Nunes and C. M. Manaia, Diversity and Antibiotic Resistance Patterns of Sphingomonadaceae Isolates from Drinking Water, Appl. Environ. Microbiol., 2011, 77, 5697–5706 CrossRef CAS PubMed.
- W. E. Blyth, E. Shahsavari, A. Aburto-Medina, A. S. Ball and A. M. Osborn, Variation in the Structure and Composition of Bacterial Communities within Drinking Water Fountains in Melbourne, Australia, Water, 2022, 14, 908 CrossRef CAS.
- L. K. Kimbell, E. L. LaMartina, A. D. Kappell, J. Huo, Y. Wang, R. J. Newton and P. J. McNamara, Cast iron drinking water pipe biofilms support diverse microbial communities containing antibiotic resistance genes, metal resistance genes, and class 1 integrons, Environ. Sci.: Water Res. Technol., 2021, 7, 584–598 RSC.
- C. Xi, Y. Zhang, C. F. Marrs, W. Ye, C. Simon, B. Foxman and J. Nriagu, Prevalence of Antibiotic Resistance in Drinking Water Treatment and Distribution Systems, Appl. Environ. Microbiol., 2009, 75, 5714–5718 CrossRef CAS PubMed.
- Y. Hu, L. Jiang, T. Zhang, L. Jin, Q. Han, D. Zhang, K. Lin and C. Cui, Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes, J. Hazard. Mater., 2018, 360, 364–372 CrossRef CAS PubMed.
- S. Jia, P. Shi, Q. Hu, B. Li, T. Zhang and X.-X. Zhang, Bacterial Community Shift Drives Antibiotic Resistance Promotion during Drinking Water Chlorination, Environ. Sci. Technol., 2015, 49, 12271–12279 CrossRef CAS PubMed.
- P. Shi, S. Jia, X.-X. Zhang, T. Zhang, S. Cheng and A. Li, Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water, Water Res., 2013, 47, 111–120 CrossRef CAS PubMed.
- B. P. Alcock, A. R. Raphenya, T. T. Y. Lau, K. K. Tsang, M. Bouchard, A. Edalatmand, W. Huynh, A.-L. V. Nguyen, A. A. Cheng, S. Liu, S. Y. Min, A. Miroshnichenko, H.-K. Tran, R. E. Werfalli, J. A. Nasir, M. Oloni, D. J. Speicher, A. Florescu, B. Singh, M. Faltyn, A. Hernandez-Koutoucheva, A. N. Sharma, E. Bordeleau, A. C. Pawlowski, H. L. Zubyk, D. Dooley, E. Griffiths, F. Maguire, G. L. Winsor, R. G. Beiko, F. S. L. Brinkman, W. W. L. Hsiao, G. V. Domselaar and A. G. McArthur, CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database, Nucleic Acids Res., 2020, 48, D517–D525 CrossRef CAS PubMed.
- M. R. Moore, M. Pryor, B. Fields, C. Lucas, M. Phelan and R. E. Besser, Introduction of Monochloramine into a Municipal Water System: Impact on Colonization of Buildings by Legionella spp, Appl. Environ. Microbiol., 2006, 72, 378–383 CrossRef CAS PubMed.
- M. B. Waak, R. M. Hozalski, C. Hallé and T. M. LaPara, Comparison of the microbiomes of two drinking water distribution systems-with and without residual chloramine disinfection, Microbiome, 2019, 7, 87–87 CrossRef PubMed.
- Z. Wang, J. Kim and Y. Seo, Influence of Bacterial Extracellular Polymeric Substances on the Formation of Carbonaceous and Nitrogenous Disinfection Byproducts, Environ. Sci. Technol., 2012, 46, 11361–11369 CrossRef CAS PubMed.
- J. Chen, W. Li, J. Zhang, W. Qi, Y. Li, S. Chen and W. Zhou, Prevalence of antibiotic resistance genes in drinking water and biofilms: The correlation with the microbial community and opportunistic pathogens, Chemosphere, 2020, 259, 127483 CrossRef CAS PubMed.
- M. B. Waak, T. M. LaPara, C. Hallé and R. M. Hozalski, Occurrence of Legionella spp. in Water-Main Biofilms from Two Drinking Water Distribution Systems, Environ. Sci. Technol., 2018, 52, 7630–7639 CrossRef CAS PubMed.
- Y.-H. Chuang, A. Szczuka and W. A. Mitch, Comparison of Toxicity-Weighted Disinfection Byproduct Concentrations in Potable Reuse Waters and Conventional Drinking Waters as a New Approach to Assessing the Quality of Advanced Treatment Train Waters, Environ. Sci. Technol., 2019, 53, 3729–3738 CrossRef CAS PubMed.
- Y.-H. Chuang and W. A. Mitch, Effect of Ozonation and Biological Activated Carbon Treatment of Wastewater Effluents on Formation of N-nitrosamines and Halogenated Disinfection Byproducts, Environ. Sci. Technol., 2017, 51, 2329–2338 CrossRef CAS PubMed.
- D. L. McCurry, K. P. Ishida, G. L. Oelker and W. A. Mitch, Reverse Osmosis Shifts Chloramine Speciation Causing Re-Formation of NDMA during Potable Reuse of Wastewater, Environ. Sci. Technol., 2017, 51, 8589–8596 CrossRef CAS PubMed.
- B. W. Stamps, M. B. Leddy, M. H. Plumlee, N. A. Hasan, R. R. Colwell and J. R. Spear, Characterization of the Microbiome at the World's Largest Potable Water Reuse Facility, Front. Microbiol., 2018, 9, 2435–2435 CrossRef PubMed.
- M. Sousi, G. Liu, S. G. Salinas-Rodriguez, L. Chen, J. Dusseldorp, P. Wessels, J. C. Schippers, M. D. Kennedy and W. van der Meer, Multi-parametric assessment of biological stability of drinking water produced from groundwater: Reverse osmosis vs. conventional treatment, Water Res., 2020, 186, 116317 CrossRef CAS PubMed.
- K. E. Fish, N. Reeves-McLaren, S. Husband and J. Boxall, Uncharted waters: the unintended impacts of residual chlorine on water quality and biofilms, npj Biofilms Microbiomes, 2020, 6, 1–12 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ew00858k |
| ‡ Authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2023 |