Chemical and molecular tools to probe biological sex differences at multiple length scales
Nicole E.
Félix Vélez†
 ab,
Rayyan M.
Gorashi†
ab,
Rayyan M.
Gorashi†
 ab and
Brian A.
Aguado
ab and
Brian A.
Aguado
 *ab
*ab
aDepartment of Bioengineering, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA. E-mail: baguado@eng.ucsd.edu
bSanford Consortium for Regenerative Medicine, 2880 Torrey Pines Scenic Drive, La Jolla, CA 92037, USA
First published on 24th August 2022
Abstract
Biological sex differences are observed at multiple different length scales and across organ systems. Gaps in knowledge remain regarding our understanding of how molecular, cellular, and environmental factors contribute to physiological sex differences. Here, we provide our perspective on how chemical and molecular tools can be leveraged to explore sex differences in biology at the molecular, intracellular, extracellular, tissue, and organ length scales. We provide examples where chemical and molecular tools were used to explore sex differences in the cardiovascular, nervous, immune, and reproductive systems. We also provide a future outlook where chemical and molecular tools can be applied to continue investigating sex differences in biology, with the ultimate goal of addressing inequities in biomedical research and approaches to clinical treatments.
Introduction
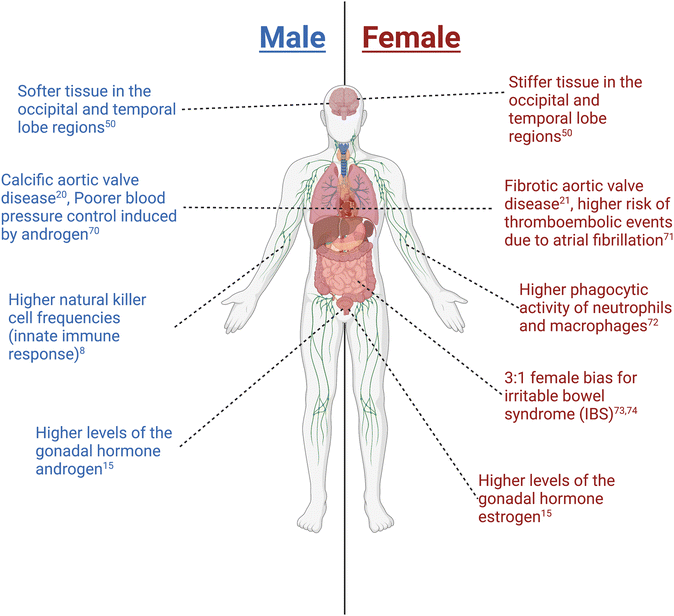
Biological sex plays a fundamental role in molecular and cellular processes during health and disease and contributes to patient responses to treatments.1,2 The term “sex” refers to biological characteristics; in humans, sex refers to attributes that distinguish female, male, and intersex individuals. Sex may be defined according to genetic sex determination (e.g., sex chromosomes), gametes/germ cell production, reproductive organs, hormones/environmental factors, or primary sex characteristics including internal reproductive organs and external genitalia. Importantly, sex attributes may or may not be aligned in any individual (e.g., external genitalia may not reflect internal genital sex). The term “gender” refers to sociocultural identities, behaviours, and attitude; the terms sex and gender are not interchangeable. Even though gender has significant implications in regulating a person's health,3 herein we will focus on the current work and future outlook on understanding biological sex differences in health and disease spanning across multiple different biological length scales.Outside of the gonads, there are organs and organ systems that exhibit sex differences (Fig. 1). First, for example, sex differences in the heart have been observed at cellular and molecular levels, where factors such as contractility of cardiac cells, electrophysiology, and gene expression have been shown to be sexually dimorphic.4 One such example is that females express genes associated with ischemic events which correlates with their higher risk of developing post-myocardial infarction relative to males.5 Second, many neurological disorders also exhibit sex differences in incidence, severity, or response to treatment. For example, the hormone estrogen provides protective effects to the blood–brain barrier, making females less susceptible to neuroinflammatory disorders relative to males.6,7 Third, on a more systemic scale, sex has been shown to affect immune responses to both self and foreign antigens, with females exhibiting higher innate immunity than males, contributing to their increased susceptibility to inflammatory and autoimmune diseases.8 Despite current efforts, the molecular and cellular mechanisms that give rise to these and other sex differences in human health and disease need to be defined to improve current therapeutic approaches for treating patients.
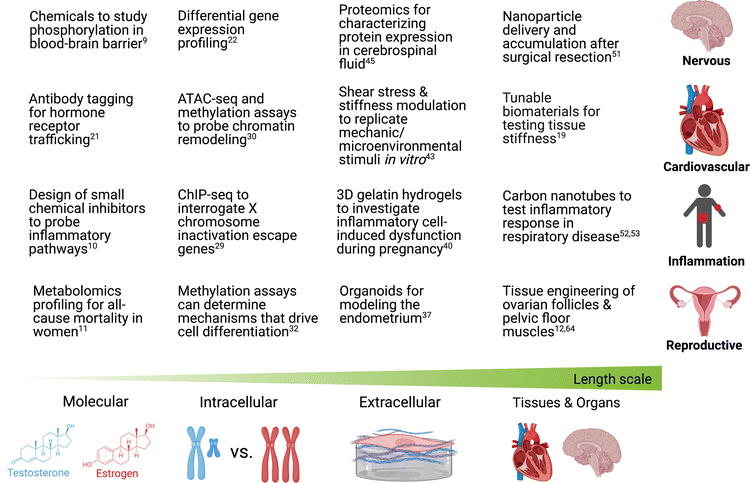
In this perspective, we outline existing chemical and molecular tools that may be used to investigate sex differences in the heart, brain, inflammation, and reproductive organs at multiple different length scales (Fig. 2). Numerous chemical tools and techniques have been developed to help probe biological processes at different length scales, yet by and large, they have rarely been exploited to study sex-specific biology. Using various chemical tools, scientists have been able to elucidate intracellular pathways, understand the relation between cells and their microenvironment, and investigate the synergy that exists within organ systems. As scientists uncover more sex differences in diseases, it will become increasingly important to use new and existing tools that can help determine biological mechanisms as a function of biological sex.9–12
Chemical tools to probe molecular sex differences
Chemical tools can be used to target specific biological macromolecules, act as effectors that alter cellular phenotypes, and provide mechanistic insights into sex-specific biological processes. Small molecule tools can also help determine localization and function of key intracellular molecules. Small molecules are defined as having a low molecular weight (between 500 Da to 1 kDa13) and are used to probe the function of individual molecules, including intracellular proteins and hormones, and identify sex differences at the smallest length scales. Some of the most common tools synthesized to probe molecular sex differences are (1) antibody tags and fluorescence resonance energy transfer (FRET) biosensors to help localize molecules of interest and (2) drugs that inhibit function of proteins or gene transcription. These techniques are coupled with optical molecular imaging techniques to help visualize molecular activity and localization within a cell.Tags and reporters
One of the most common techniques to label intracellular molecules is antibody tagging. With antibody tagging, molecules of interest can be isolated or visualized to understand signaling activity within cells. The synthesis of highly-specific antibody probes at the near infrared spectra has been a useful tool in studying atherosclerosis to identify intravascular lesion sites of coronary artery disease.14 Since atherosclerosis has well-known but poorly understood sexual dimorphisms associated with differing levels of testosterone and the presence of the Y chromosome,15 this tool would prove useful in probing the mechanisms that cause increased plaque burden and inflammation observed in males relative to females with vascular disease. Additionally, the ability to generate various isoforms of antibody receptors has aided in studying hormone trafficking within cardiac myocytes.16 Sex hormones are strongly correlated with cardiovascular health in males and females. For example, the presence of estrogen receptors (ERs) in the heart confers protection of cardiovascular disease while loss of ERs leads to increased risk of developing heart disease, as seen in post-menopausal women.16 Synthesizing fluorescent antibody tags for the different isoforms of estrogen receptor-α (ERα) ERα66, ERα46, and ERα36 has shown the importance of ERα66 in transcriptional activation of cardiac myocytes and clarified conflicting reports of ERα activity in the MAPK or PI3K signaling pathway.16FRET-based calcium biosensors can measure changes in calcium concentration by quantifying changes in receptor domain conformation once the ion is bound to the calcium receptor.17 FRET biosensors has been used to monitor and study cell activation patterns in neurons, T lymphocytes, and myofibroblasts.17,18 Batan et al. reported the use of FRET-based calcium sensors to test the mechanosensitive role of Transient Receptor Potential Vanilloid type 4 (TRVP4) during myofibroblast activation in aortic valvular interstitial cells (VICs).18 The authors reported that TRVP4 partly regulates myofibroblast activation via Yes-associated protein (YAP) nuclear localization induction and identified this mechanism as a potential therapeutic target for valve fibrosis. Since valve fibrosis is sexually dimorphic, FRET-based calcium sensors may be particularly useful to investigate sex dimorphisms in calcium shuttling in contractile cardiomyocytes and other cardiac cells.19,20
Chemical inhibition
Small molecule drugs used to inhibit signaling pathways are valuable chemical tools to elucidate mechanisms that give rise to sex-specific cellular phenotypes by blocking expression or function of messengers or receptors. For example, differential gene expression is associated to sex differences in neuronal activity in the brain, which often precedes the development of neurodegenerative diseases.21 Previous work has mapped the differential gene expression within the MAPK/ERK pathway in neurons due to different induced activity patterns.22 Here, they suppressed MAPK/ERK signaling using NMDA blockers and found that neuronal activity patterns are dependent on activity-regulated genes within this pathway. Sex differences of MAPK expression have been observed previously, with female mice showing lower levels of MAPK relative to males,23 indicating that chemical inhibitors may be useful tools for probing sex-specific signaling pathway activity.RhoA/ROCK signaling, a mechanosensitive pathway that regulates actin cytoskeletal formation and is associated with several cardiovascular diseases, is uniquely enriched in male fibroblast-like valvular interstitial cells (VICs).19 RhoA/ROCK activation partly regulates the fibroblast-to-myofibroblast transition in VICs and inhibition of its signaling using RhoA/ROCK inhibitors (Y-27632 and H-1152) exhibits sex differences, with female myofibroblasts having a decreased response relative to males due to increased expression of genes that escape X-chromosome inactivation in female myofibroblasts.19
Additionally, using inhibitors to block lipoxygenase formation has been useful in understanding the role signaling molecules obtained by the oxidation of unsaturated lipids, such as eicosanoids or prostaglandins, in the sex-dependent incidence of inflammation in immune diseases. Previous work using lipoxygenase inhibitors demonstrated that increased production of prostaglandins in males leads to increased levels of nuclear factor-kappa B activation, contributing to pathologies such as sepsis and post-surgery infections which display a higher incidence in males.24
Chemical tools to probe intracellular sex differences
Chemical tools used to probe intracellular molecular mechanisms can be used to evaluate sex differences in genetic and epigenetic activity between cells. Although sex disparities are often attributed to the actions of sex hormones, increasing evidence demonstrates that sex chromosomes also contribute to sex differences independently of gonads, affecting different organs and biological processes such as cardiovascular disease and metabolism.25–27 The chemical tools to probe intracellular mechanisms described here consist of chemical assays or methods that can analyze protein interactions within the cell membrane, elucidating how sex can alter intracellular activity. Genomic tools can also help probe the regulation of gene expression via epigenetic modifications. Specifically, we outline methods that can be used to understand how sex chromosomes are responsible for the sex dimorphisms observed at a cellular level.Epigenomic tools
Epigenetic mechanisms are important regulators of cell identity, and chromatin structure is considered a developmental cell type-specific filter of genomic sequence information and expression.28 Moreover, epigenetics of cells have been shown to be key regulators of sex differences in the brain, such as increased methylation in developing females brains allowing for increased synaptic functions.6 The Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) is a valuable tool that can be used to interrogate the overall epigenetic profile of a cell population. Since the study of epigenetic sex differences is a new field, ATAC-seq can be implemented to identify changes in chromatin accessibility based on cell sex.Other, more specific, genomic tools can complement ATAC-seq to identify specific transcription factors and genes that are differentially expressed between males and females. For example, chromatin immunoprecipitation (ChIP) assays can help identify transcription factors or DNA binding domains of interest associated with the different chromatin structure observed across the whole genome. ChIP has been useful in understanding how demethylation of histone H3K27 induced by the expression of X chromosome inactivation escape genes kdm5c and kdm6a caused upregulation of inflammatory interferon regulatory factors (IRFs) in microglia.29 The sexually dimorphic upregulation of IRFs contributes to the activation of microglia during post-stroke neuroinflammation and plays a role in secondary neuronal damage caused by a persistent immune response from microglia.29
Histone modifications have been shown to play an important role in fibroblast activation and fibrogenesis in the heart, lungs, and kidneys.30,31 Thus, evaluating the activity of histone modifiers can provide more information on how sex can drive epigenetic alterations. Histone deacetylase (HDAC) fluorometric assays performed on fibroblasts of the aortic heart valves have revealed increased deacetylation in pathogenically activated myofibroblasts relative to healthy fibroblasts.30 Similarly, DNA methyltransferase assays are used to evaluate the epigenetic landscape of the developing brain, demonstrating that, in developing mammals, masculinization is induced by decreased DNA methylation of the preoptic area of the brain.32
Transcriptomic tools
Transcriptome sequencing, or RNA-seq, can provide data on the transcriptionally active elements within genomes.33 In aortic valve stenosis (AVS), RNA-seq was used to evaluate valvular myofibroblast activation pathways in male and female porcine valvular interstitial cells.19 Aguado et al. observed differential gene expression between male and female myofibroblasts and identified signaling pathways that contribute to sex dimorphisms in AVS. In a similar form, using global transcriptome sequencing in T cells of mice revealed a sex dimorphism associated with increased autoimmune susceptibility in females relative to males.34RNA-seq coupled with fluorescence in situ hybridization (FISH) can help localize transcriptomic data onto the epigenetic landscape. This is especially useful for tracking transgenes created to separate sex hormones from sex chromosomes, as reported by Itoh et al. Having created a mouse model with a deletion of the Sry gene from the Y chromosome and expressed and Sry transgene in an autosome, the authors were able to identify the location and number of copies of the transgene.35 Thus, transcriptome sequencing in conjunction with visualization techniques and chemical probes can be a powerful tool in mapping sexually dimorphic intercellular activity.
Chemical tools to probe extracellular sex differences
Chemical tools can also be utilized to gain knowledge about how extracellular factors impact sex-specific biological mechanisms within cells. These mechanisms include extracellular signaling molecules that can influence signal transduction pathways and direct short-term cell response through gene activity regulation. Biomaterials can be leveraged to model microenvironments with physiologically relevant biological and mechanical properties. Proteomic techniques allow us to identify proteins of interest that could be upregulated due to specific signal transduction pathways. Together, biomaterial and proteomic tools have contributed to our collective progress in the study of sex differences.Biomaterial tools
Biomaterials are versatile tools where different chemistries may used to synthesize or modify materials, which can significantly alter their mechanical and biological properties.36 When evaluating sex differences in the extracellular space, polyethylene glycol (PEG)-based hydrogels and gelatin hydrogels have most commonly been employed as chemical tools to recapitulate key aspects of the extracellular matrix.At the extracellular level, female reproductive health has been studied using both PEG and gelatin hydrogels. 3D PEG-based hydrogels have been leveraged as tools to culture endometrial organoids to study dynamic processes involved in growth, differentiation, and breakdown due to fluctuations in sex hormone levels during endometriosis.37 Endometriosis is defined as endometrial tissue growth in the peritoneal region outside of the uterus.38 Of particular interest is the dependency of estrogen and resistance of progesterone on the pathology of the disease, leading to chronic inflammation, pain, and infertility.39 Sex hormone levels have been implicated in classification and severity of immune response.8 In their model, epithelial–stromal cell interactions were studied as crosstalk between these cells drives the endometrial response to sex hormones. PEG vinyl sulfone (PEG-VS) gels were designed using fibronectin-derived peptides (e.g., RGD), collagen-I derived peptides (e.g. GFOGER), peptides with affinities for fibronectin and collagen IV, and a dithiol crosslinking peptide with a matrix metalloproteinase (MMP)-sensitive sequence. All of the conjugated peptides represent proteins integral to the development and maintenance of the extracellular matrix (ECM) in the endometrium. By tuning the hydrogel stiffness to that of native endometrial tissue, the hydrogel-cell constructs mimicked menstrual specific reproductive events.
3D biomaterials have also been used to investigate inflammation- and stromal cell-induced dysregulation of epithelial proliferation. Methacrylamide-functionalized gelatin (GelMA) hydrogels were employed to delineate the signaling pathway during trophoblast invasion for the establishment of pregnancy.40 In particular, GelMA provided the hydrogel network necessary to study the motility of trophoblasts and the signaling mechanisms of pregnancy-specific glycoproteins (PSG) such as PSG9 and PSG1. They identified PSG9 and PSG1 as key regulators of trophoblast motility where PSG9 reduces motility and PSG1 increases motility. The encapsulated spheroids used in this study provided the researchers with the tool necessary to understand trophoblast motility and remodeling response in the presence and absence of soluble factors and extracellular matrix cues.
In the study of aortic valve stenosis (AVS), 3D PEG-based hydrogels with conjugated peptides have been used to characterize valve cell interactions, particularly that of valvular interstitial cells (VICs) and valvular endothelial cells (VECs).41 Given that AVS is a sexually dimorphic disease,20 and VICs have been shown to have sexually dimorphic transcriptomes,19,42 advanced biomaterial strategies to enable co-culture of VICs with other cell types would be desirable for further probing sex-specific cellular behaviours. In this study, arginine–lysine–arginine (RKR) and arginine–glycine–aspartate (RGD) were incorporated into the PEG hydrogel. RKR is a laminin-derived syndecan binding peptide sequence and RGD is a fibronectin-derived integrin binding peptide sequence, both of which can support valve cell adhesion to the hydrogel matrix. 3D PEG-based hydrogel models of AVS allow for the spatial arrangement of VICs and VECs to mimic that of an in situ aortic valve. Homeostatic functions were maintained using this model and VEC monolayer formation was introduced using both RKR and RGD binding peptides. The next generation in vitro models of VIC–VEC interactions may involve sex-separation of these cell types to investigate potential sex differences in co-culture environments.
In other cardiovascular applications, modulation of shear stress (0.5–1.5 Pa) and substrate stiffness (10–100 kPa) has been shown to result in sexual dimorphisms human umbilical vein endothelial cell (HUVEC) behavior.43 This study was conducted in the previously developed MechanoBioTester system that includes a chamber with a tunable polydimethylsiloxane (PDMS) elastomer substrate for cell culture.44 To facilitate cell adhesion, the PDMS substrate was coated with collagen I. In this study, yes-associated protein 1 (YAP-1) nuclear localization was shown to increase in male HUVECs in response to the combination of increasing shear stress and substrate stiffness. In female HUVECs, however, YAP-1 nuclear localization was not significantly changed in response to the same conditions. Collectively, multiple studies suggest several different cell types exhibit sexually dimorphic responses to engineered microenvironments, suggesting biomaterials may serve as useful tools for recapitulating sex differences in cellular responses to fluid flow in vivo.
Proteomic tools
Proteomics involves the analysis of protein expression in a target microenvironment, such as serum, cerebrospinal fluid, urine, or cell lysates. Proteomic tools are used to compare protein abundances in different samples using mass spectrometry methods.45 An especially relevant proteomic tool to study sex-specific biology is the SOMAscan DNA aptamer array (SomaLogic). The basis of the technology for the SOMAscan array lies in its chemically modified SOMAmers, which are synthetic molecular recognition elements. These SOMAmers have functional groups that allow for high-affinity interaction with specific protein surfaces. The key innovation in the SOMAmers is the wide variety of chemical modifications that allow for a library of over 7000 proteins to be identified within the target fluid. Additionally, the selection of SOMAmers with slow dissociation rates ensures a higher affinity through the disruption of nonspecific binding interactions.46In the study of AVS, SOMAscan DNA aptamer arrays have been employed to develop a sex-specific profile of serum factors before and after a transcatheter aortic valve replacement (TAVR).47 TAVR is the current standard of care for surgical replacement of the aortic valve in patients with severe AVS. Sex differences in pre and post TAVR serum factors were identified using SomaLogic proteomic analysis techniques. In males, 347 proteins had altered changes in abundance, whereas only 39 proteins had abundance changes in females. Through the use of patient-specific sera and physiologically relevant biomaterials-based models of the valve ECM, sex-specific outcomes in post-TAVR cardiac remodeling observed in vivo (e.g., males experience more improved left ventricular remodeling after TAVR relative to females) were recapitulated in vitro.
Chemical tools to probe tissue/organ scale sex differences
Chemical tools can also provide insight into biological mechanisms that guide health and disease involving the whole tissue or organ. These mechanisms involve elements such as varying cell types, extracellular matrix proteins, cytokines, and their interactions with the tissue. Several groups have leveraged chemical tools as a means to investigate tissue responses to innovations such as nanomaterials, biomaterials-based vaccines, and biomaterials for sex-specific tissue regeneration. Additionally, existing chemical tools can also be repurposed in future studies to investigate sex-specific pathology in tissue and organ behavior.Nanomaterials
Nanomaterials carry unique physicochemical properties, such as shape, size, porosity, and surface charge, that can be optimized for different tissue microenvironments.48 Nanomaterials tools can also be composed of polystyrene, liposomes, and metals,49 are on a size range of 1–100 nm, and may take shapes such as spheres, rods, and tubes.48 Sex-based differences at the tissue scale have been observed through the utilization of these nanomaterials in many in vivo studies, from drug delivery to disease modeling.In the brain, sex differences have been observed in brain tissue stiffness across different regions, such as the temporal and occipital lobes.50 The differences in stiffness are predicted to be 0.23 kPa and 0.09 kPa between females and age-matched males. For procedures involving surgical interventions of brain tissue, such as surgical resection, nanoparticles can be utilized as a potential therapeutic delivery vehicle. However, nanoparticle (NP) deposition to peri-resection brain tissue following surgical resection is not well-characterized. An in vivo study delivered polystyrene NPs (PS-NPs) with surface chemical modifications intravenously to the brain of a mouse.51 Here, a resection model to mimic surgical injury was developed where C57BL/6 mice had a section of their frontal lobe physically removed by a vacuum apparatus. Though the exact mechanism of adhesion remains unclear, PS-NPs were found to be present only in the vasculature of the brain. The chemical modifications for this NP system included N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide to allow for the covalent attachment of methoxy-PEG-amine to the surface of PS-NPs. These chemical modifications facilitate binding of the PS-NPs to the surface of cells to ultimately increase uptake in the tissue. In this study, the NP deposition profile for males did not have temporal dependence.
In contrast, another study used PS-NPs with similar carbodiimide surface chemistry in a traumatic brain injury (TBI) model and observed sex-dependent increases in nanoparticle accumulation in males, but not females. Males were observed to have temporal dependence for the NP deposition profile, where a secondary uptake peak could be observed days after the initial injection.52 While the studies employed nearly identical PS-NPs, they each yielded different results. This observance underscores the need to expand upon these results and study PS-NP uptake in varying regions of the brain tissue. As such, parameters such as regional brain tissue stiffness, an established sexual dimorphism, need to be accounted for in experimental design.
Some groups have investigated sex differences in inflammatory responses using nanomaterials to induce respiratory disease in in vivo mouse models. Nickel NPs can be used to mimic respiratory disease as a result of chronic nickel exposure from an occupational hazard. C57BL/6 mice were exposed to nickel NPs via oropharyngeal aspiration either once (acute exposure) or over a twenty-day time frame (subchronic exposure).53 Sex differences in both the acute and subchronic timepoints were observed. After acute exposure, males experienced an enhanced inflammatory status as determined by elevated inflammatory chemokines (C–X–C motif ligand 1, CXCL1) and cytokines (interleukin-6, IL6). In addition, a higher population of neutrophils was present in the bronchoalveolar lavage fluid (BALF).
After subchronic exposure to nickel NPs, lung tissue in female mice was found to have increased expression of signal transducer and activator of transcription 1, STAT1. Another study investigated sex bias for the development of respiratory diseases using multiwalled carbon walled nanotubes (MWCNTs) and crystalline silica (cSiO2).54 MWCNTs had a 27 nm diameter, 5–15 μm length, and made up of 5.54% of nickel.55 After acute exposure to MWCNTs, female mice experienced a greater eosinophilic response and increased type 2 inflammatory cytokine production in the lung lavage fluid relative to males. In contrast to females, subchronic exposure to cSiO2 NPs resulted in severe alveolitis accompanied with an increased presence of dendritic cells. These studies leveraged nanomaterials to induce respiratory disease and examine sex dimorphisms in inflammatory responses. Future applications of these nanomaterials can delineate roles of inflammatory chemokines and cytokines between the sexes and investigate the influence of long-term pathology on immune responses.
Biomaterials-based vaccine delivery and immune response
Biomaterials with different surface chemistries are useful for modulating immune responses and inducing immunity to pathogens. Common biomaterials for vaccine delivery include lipid-based delivery vehicles, natural polymer-based particles, and synthetic polymer-based particles.56 Sex differences in vaccine delivery come into play in the immune response to the foreign biomaterials. For example, sex differences in innate immunity can be attributed to Toll-like receptor 7 (TLR7) gene escaping X chromosome inactivation.57 Elevated expression of TLR7 in conjunction with increased interferon-gamma production results in a higher innate immune response in females relative to males. Increased interferon-gamma production can be attributed to the downstream effect of TLR7 on increased inflammatory cytokine production.58 One study cited androgen response elements and oestrogen response elements as key promoters in genes involved in the innate immune response, leading to a potential mediator in sex dimorphisms.59Sex differences in outcomes of the novel coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have also been identified. Male bias in the mortality of the ongoing COVID-19 pandemic has been observed worldwide.60–62 In fact, sex differences in COVID-19 pathogenesis and mortality is congruent with other viral infections. One group utilized a natural polymer-based particle to study vaccine uptake in an in vivo model of SARS-CoV-2.63 The natural polymer-based vehicle for COVID-19 vaccine was fabricated through a redox-induced free radical cryo-polymerization of hyaluronic acid glycidyl methacrylate (HAGM). The HAGM cryogel vehicle containing the COVID-19 vaccine was then injected into the mouse along with a booster dose 3 weeks after. Results were promising as immunomodulatory effects of the HAGM cryogel led to B cell stimulation and recruitment as well as CD4+ and CD8+ T cell responses. A major limitation of this study, given prior knowledge of immune responses, is the lack of sex disaggregation in data collection. Future work seeking to investigate biomaterial-based vaccine delivery and immune response must consider sex as a biological variable.
Biomaterials for tissue regeneration and drug delivery
While we previously discussed biomaterials as a tool for the investigation of extracellular mechanisms, we can also consider the injection or implantation of biomaterials as a tool for local tissue regeneration. Such biomaterials can be prepared hydrogels or electrospun fibers for a variety of women's reproductive health applications.64Pelvic floor muscle dysfunction can often be attributed to muscle atrophy and the development of fibrosis from an activated inflammatory state.65 To mediate these dysfunctions, a hydrogel comprised of skeletal muscle extracellular matrix (SKM) was used in an in vivo model of simulated birth injury in rats. The SKM hydrogel was fabricated from the decellularized longissimus dorsi muscle of pig and underwent characterization to measure protein fragment molecular weights through liquid chromatography with tandem mass spectrometry. SKM hydrogel injection following simulated birth injury decreased collagen deposition in the pelvic floor muscle tissue.65 This study exemplifies the therapeutic potential of hydrogels composed of decellularized extracellular matrix. Future applications can leverage the decellularization technique to promote regeneration of other tissue within the female reproductive tract.
Electrospun fibers have also been utilized as a tool for sexually transmitted disease prevention, contraception, and treatment of infection. These fibers are prepared from polymer solutions that are subjected to an electric field and thus pulled into a fiber shape.66 As with other biomaterials discussed earlier, properties such as porosity, stiffness, and size can still be tuned for the target tissue microenvironment. In the reproductive tract, electrospun fibers have been applied in the treatment of bacterial vaginosis and fungal candidiasis (yeast infections). Mucoadhesive electrospun fibers composed of polyvinyl alcohol were loaded with Fluconazole, a common drug for the treatment of yeast infections. These fibers were able to selectively target Candida albicans fungal population in the treatment of candidiasis.67 The group found that their fibers ranged between 150–180 nm and allowed for sustained drug release of Fluconazole over a period of 6 hours. The advancement of mucoadhesion extends the application of this biomaterial for female reproductive tract applications, improving efficacy of drug delivery. Future researchers can incorporate this property into their studies to achieve similar success.
Conclusions and future outlook
Incorporating the chemical tools outlined in this perspective into sex differences research, particularly in a pathophysiological context, will aid in bridging the gap in current health disparities that affect the female population.68 A thorough understanding of the pathways and mechanisms involved in sex dimorphisms would enable the development of targeted and improved therapies as a function of sex. At the molecular level, the use of antibody tags, FRET biosensors, optical molecular imaging, and chemical inhibition can all be used to help illuminate pathways and construct sex-specific networks of cell types across different tissues. Considering the ubiquitous presence of sex hormones around the body, it is important to consider sex hormone pathways and how these can affect transcriptional activity in cell types across the body. Understanding at the smallest scale how male and female cells regulate their processes provides insight for more specific and effective therapeutic targets. At a larger scale, precision biomaterials,69 proteomics, and nanomaterials allow us to understand the extracellular mechanisms that lead to sex dimorphisms in disease. Each of these chemical tools can elucidate cell–cell, cell–tissue, and tissue–organ system interactions that regulate sex differences in biology. Although we highlight important advances in using chemical and molecular tools to study sexual dimorphisms, opportunities exist to investigate additional biological features (e.g., hemodynamics) that have not yet been explored using these tools. As sex-specific mechanisms are better characterized using various chemical and molecular tools, we posit the development of future sex-based therapies will help achieve improved treatment outcomes for patients of different biological sex.Author contributions
N. E. F. V., R. M. G., and B. A. A. conceptualized the article, wrote the original draft, and reviewed/edited the article. B. A. A. served as project administrator.Conflicts of interest
There are no conflicts to declare.Acknowledgements
B. A. A. acknowledges funding from the National Institutes of Health (R00 HL145817 and R25 HL145817) and the American Heart Association (942253).70–74References
- C. J. Walker, M. E. Schroeder, B. A. Aguado, K. S. Anseth and L. A. Leinwand, J. Mol. Cell. Cardiol., 2021, 160, 42–55 CrossRef CAS PubMed.
- N. I. o. H. (NIH), NIH, Notice Number NOT-OD-15-102.
- P. Sanchez-Lopez Mdel, I. Cuellar-Flores and V. Dresch, Women Health, 2012, 52, 182–196 CrossRef PubMed.
- G. T. Squiers, M. A. McLellan, A. Ilinykh, J. Branca, N. A. Rosenthal and A. R. Pinto, Cardiovasc. Res., 2021, 117, 2252–2262 CrossRef CAS PubMed.
- G. Stone, A. Choi, M. Oliva, J. Gorham, M. Heydarpour, C. E. Seidman, J. G. Seidman, S. F. Aranki, S. C. Body, V. J. Carey, B. A. Raby, B. E. Stranger and J. D. Muehlschlegel, Hum. Mol. Genet., 2019, 28, 1682–1693 CrossRef CAS PubMed.
- E. Y. Shen, T. H. Ahern, I. Cheung, J. Straubhaar, A. Dincer, I. Houston, G. J. de Vries, S. Akbarian and N. G. Forger, Exp. Neurol., 2015, 268, 21–29 CrossRef CAS PubMed.
- E. Maggioli, S. McArthur, C. Mauro, J. Kieswich, D. H. M. Kusters, C. P. M. Reutelingsperger, M. Yaqoob and E. Solito, Brain, Behav., Immun., 2016, 51, 212–222 CrossRef CAS PubMed.
- S. L. Klein and K. L. Flanagan, Nat. Rev. Immunol., 2016, 16, 626–638 CrossRef CAS PubMed.
- Y. Hashimoto and M. Campbell, Biochim. Biophys. Acta, Biomembr., 2020, 1862, 183298 CrossRef CAS PubMed.
- X. Ren, X. Li, L. Jia, D. Chen, H. Hou, L. Rui, Y. Zhao and Z. Chen, FASEB J., 2017, 31, 711–718 CrossRef CAS PubMed.
- R. Balasubramanian, N. P. Paynter, F. Giulianini, J. E. Manson, Y. Zhao, J. C. Chen, M. Z. Vitolins, C. A. Albert, C. Clish and K. M. Rexrode, Int. J. Epidemiol., 2020, 49, 289–300 CrossRef PubMed.
- P. D. Rios, E. Kniazeva, H. C. Lee, S. Xiao, R. S. Oakes, E. Saito, J. S. Jeruss, A. Shikanov, T. K. Woodruff and L. D. Shea, Biotechnol. Bioeng., 2018, 115, 2075–2086 CrossRef CAS PubMed.
- P. Fechner, O. Bleher, M. Ewald, K. Freudenberger, D. Furin, U. Hilbig, F. Kolarov, K. Krieg, L. Leidner, G. Markovic, G. Proll, F. Pröll, S. Rau, J. Riedt, B. Schwarz, P. Weber and J. Widmaier, Anal. Bioanal. Chem., 2014, 406, 4033–4051 CrossRef CAS PubMed.
- R. Y. Khamis, K. J. Woollard, G. D. Hyde, J. J. Boyle, C. Bicknell, S.-H. Chang, T. H. Malik, T. Hara, A. Mauskapf, D. W. Granger, J. L. Johnson, V. Ntziachristos, P. M. Matthews, F. A. Jaffer and D. O. Haskard, Sci. Rep., 2016, 6, 21785 CrossRef CAS PubMed.
- A. P. Arnold, L. A. Cassis, M. Eghbali, K. Reue and K. Sandberg, Arterioscler., Thromb., Vasc. Biol., 2017, 37, 746–756 CrossRef CAS PubMed.
- E. K. Pugach, C. L. Blenck, J. M. Dragavon, S. J. Langer and L. A. Leinwand, Mol. Cell. Endocrinol., 2016, 431, 62–70 CrossRef CAS PubMed.
- T. Thestrup, J. Litzlbauer, I. Bartholomäus, M. Mues, L. Russo, H. Dana, Y. Kovalchuk, Y. Liang, G. Kalamakis, Y. Laukat, S. Becker, G. Witte, A. Geiger, T. Allen, L. C. Rome, T.-W. Chen, D. S. Kim, O. Garaschuk, C. Griesinger and O. Griesbeck, Nat. Methods, 2014, 11, 175–182 CrossRef CAS PubMed.
- D. Batan, D. K. Peters, M. E. Schroeder, B. A. Aguado, M. W. Young, R. M. Weiss and K. S. Anseth, FASEB J., 2022, 36, e22306 CrossRef CAS PubMed.
- B. A. Aguado, C. J. Walker, J. C. Grim, M. E. Schroeder, D. Batan, B. J. Vogt, A. G. Rodriguez, J. A. Schwisow, K. S. Moulton, R. M. Weiss, D. D. Heistad, L. A. Leinwand and K. S. Anseth, Circulation, 2022, 145, 513–530 CrossRef CAS PubMed.
- L. Simard, N. Cote, F. Dagenais, P. Mathieu, C. Couture, S. Trahan, Y. Bosse, S. Mohammadi, S. Page, P. Joubert and M. A. Clavel, Circ. Res., 2017, 120, 681–691 CrossRef CAS PubMed.
- S. Fernández-García, J. G. Orlandi, G. A. García-Díaz Barriga, M. J. Rodríguez, M. Masana, J. Soriano and J. Alberch, BMC Biol., 2020, 18, 58 CrossRef PubMed.
- K. M. Tyssowski, N. R. Destefino, J.-H. Cho, C. J. Dunn, R. G. Poston, C. E. Carty, R. D. Jones, S. M. Chang, P. Romeo, M. K. Wurzelmann, J. M. Ward, M. L. Andermann, R. N. Saha, S. M. Dudek and J. M. Gray, Neuron, 2018, 98, 530–546 CrossRef CAS PubMed.
- J.-T. Yang, Z.-J. Wang, H.-Y. Cai, L. Yuan, M.-M. Hu, M.-N. Wu and J.-S. Qi, Neurosci. Bull., 2018, 34, 736–746 CrossRef CAS PubMed.
- S. Pace, A. Rossi, V. Krauth, F. Dehm, F. Troisi, R. Bilancia, C. Weinigel, S. Rummler, O. Werz and L. Sautebin, Sci. Rep., 2017, 7, 3759 CrossRef PubMed.
- A. P. Arnold and X. Chen, Front. Neuroendocrinol., 2009, 30, 1–9 CrossRef PubMed.
- J. C. Link, C. B. Wiese, X. Chen, R. Avetisyan, E. Ronquillo, F. Ma, X. Guo, J. Yao, M. Allison, Y.-D. I. Chen, J. I. Rotter, J. S. El-Sayed Moustafa, K. S. Small, S. Iwase, M. Pellegrini, L. Vergnes, A. P. Arnold and K. Reue, J. Clin. Invest., 2020, 130, 5688–5702 CrossRef CAS PubMed.
- W. Shi, X. Sheng, K. M. Dorr, J. E. Hutton, J. I. Emerson, H. A. Davies, T. D. Andrade, L. K. Wasson, T. M. Greco, Y. Hashimoto, J. D. Federspiel, Z. L. Robbe, X. Chen, A. P. Arnold, I. M. Cristea and F. L. Conlon, Dev. Cell, 2021, 56, 3019–3034 CrossRef CAS PubMed.
- M. Perino and G. J. C. Veenstra, Dev. Cell, 2016, 38, 610–620 CrossRef CAS PubMed.
- S. Qi, A. Al Mamun, C. Ngwa, S. Romana, R. Ritzel, A. P. Arnold, L. D. McCullough and F. Liu, J. Neuroinflammation, 2021, 18, 70 CrossRef CAS PubMed.
- C. J. Walker, C. Crocini, D. Ramirez, A. R. Killaars, J. C. Grim, B. A. Aguado, K. Clark, M. A. Allen, R. D. Dowell, L. A. Leinwand and K. S. Anseth, Nat. Biomed. Eng., 2021, 5, 1517–1518 CrossRef PubMed.
- S. K. Huang, A. M. Scruggs, J. Donaghy, J. C. Horowitz, Z. Zaslona, S. Przybranowski, E. S. White and M. Peters-Golden, Cell Death Dis., 2013, 4, e621 CrossRef CAS.
- B. M. Nugent, C. L. Wright, A. C. Shetty, G. E. Hodes, K. M. Lenz, A. Mahurkar, S. J. Russo, S. E. Devine and M. M. McCarthy, Nat. Neurosci., 2015, 18, 690–697 CrossRef CAS PubMed.
- P. De Wit, M. H. Pespeni, J. T. Ladner, D. J. Barshis, F. Seneca, H. Jaris, N. O. Therkildsen, M. Morikawa and S. R. Palumbi, Mol. Ecol. Resour., 2012, 12, 1058–1067 CrossRef CAS PubMed.
- Y. Itoh, L. C. Golden, N. Itoh, M. A. Matsukawa, E. Ren, V. Tse, A. P. Arnold and R. R. Voskuhl, J. Clin. Invest., 2019, 129, 3852–3863 CrossRef PubMed.
- Y. Itoh, R. Mackie, K. Kampf, S. Domadia, J. D. Brown, R. O’Neill and A. P. Arnold, BMC Res. Notes, 2015, 8, 69 CrossRef PubMed.
- M. Los, A. Hudecki and E. Wiecheć, Stem cells and biomaterials for regenerative medicine, Academic Press, an imprint of Elsevier, London, United Kingdom; San Diego, CA, 2019 Search PubMed.
- A. B. Juan, S. Gnecco, K. Buttrey, C. Ives, B. Goods and V. H.-G. Lauren Baugh, M. Loring, K. Issacson and L. G. Griffith, 2021, bioRxiv DOI:10.1101/2021.09.30.462577.
- P. R. Koninckx, A. Ussia, L. Adamyan, A. Wattiez and J. Donnez, Fertil. Steril., 2012, 98, 564–571 CrossRef PubMed.
- E. Garcia-Gomez, E. R. Vazquez-Martinez, C. Reyes-Mayoral, O. P. Cruz-Orozco, I. Camacho-Arroyo and M. Cerbon, Front. Endocrinol., 2019, 10, 935 CrossRef.
- S. G. Zambuto, S. Rattila, G. Dveksler and B. A. C. Harley, Cell. Mol. Bioeng., 2022, 15, 175–191 CrossRef CAS PubMed.
- D. S. Puperi, L. R. Balaoing, R. W. O'Connell, J. L. West and K. J. Grande-Allen, Biomaterials, 2015, 67, 354–364 CrossRef CAS PubMed.
- C. M. McCoy, D. Q. Nicholas and K. S. Masters, PLoS One, 2012, 7, e39980 CrossRef CAS PubMed.
- B. D. James and J. B. Allen, Adv. Healthcare Mater., 2021, 10, e2100735 CrossRef PubMed.
- B. D. James, N. Montoya and J. Allen, ACS Biomater. Sci. Eng., 2020, 6, 3673–3689 CrossRef CAS PubMed.
- R. Zhang, L. Barker, D. Pinchev, J. Marshall, M. Rasamoelisolo, C. Smith, P. Kupchak, I. Kireeva, L. Ingratta and G. Jackowski, Proteomics, 2004, 4, 244–256 CrossRef CAS PubMed.
- L. Gold, J. J. Walker, S. K. Wilcox and S. Williams, Nat. Biotechnol., 2012, 29, 543–549 CAS.
- B. A. Aguado, K. B. Schuetze, J. C. Grim, C. J. Walker, A. C. Cox, T. L. Ceccato, A. C. Tan, C. C. Sucharov, L. A. Leinwand, M. R. G. Taylor, T. A. McKinsey and K. S. Anseth, Sci. Transl. Med., 2019, 11, eaav3233 CrossRef CAS PubMed.
- M. M. Modena, B. Ruhle, T. P. Burg and S. Wuttke, Adv. Mater., 2019, 31, e1901556 CrossRef PubMed.
- M. Colombo, S. Carregal-Romero, M. F. Casula, L. Gutierrez, M. P. Morales, I. B. Bohm, J. T. Heverhagen, D. Prosperi and W. J. Parak, Chem. Soc. Rev., 2012, 41, 4306–4334 RSC.
- A. Arani, M. C. Murphy, K. J. Glaser, A. Manduca, D. S. Lake, S. A. Kruse, C. R. Jack, Jr., R. L. Ehman and J. Huston, 3rd, NeuroImage, 2015, 111, 59–64 CrossRef PubMed.
- O. B. Sara Belko, C. Baker, M. Fowler, M. P. Masouleh, K. T. Householder, S. E. Stabenfeldt and R. W. Sirianni, Res. Square, 2021 DOI:10.21203/rs.3.rs-704464/v1.
- V. N. Bharadwaj, C. Copeland, E. Mathew, J. Newbern, T. R. Anderson, J. Lifshitz, V. D. Kodibagkar and S. E. Stabenfeldt, Tissue Eng., Part A, 2020, 26, 688–701 CrossRef CAS PubMed.
- D. J. You, H. Y. Lee, A. J. Taylor-Just, K. E. Linder and J. C. Bonner, Nanotoxicology, 2020, 14, 1058–1081 CrossRef CAS PubMed.
- J. L. Ray and A. Holian, Inhal Toxicol, 2019, 31, 285–297 CrossRef CAS PubMed.
- R. F. Hamilton, Jr., C. Xiang, M. Li, I. Ka, F. Yang, D. Ma, D. W. Porter, N. Wu and A. Holian, Inhal. Toxicol., 2013, 25, 199–210 CrossRef PubMed.
- P. Sahdev, L. J. Ochyl and J. J. Moon, Pharm. Res., 2014, 31, 2563–2582 CrossRef CAS PubMed.
- P. Pisitkun, J. A. Deane, M. J. Difilippantonio, T. Tarasenko, A. B. Satterthwaite and S. Bolland, Science, 2006, 312, 1669–1672 CrossRef CAS PubMed.
- T. Takahashi and A. Iwasaki, Science, 2021, 371, 347–348 CrossRef CAS.
- M. F. Hannah, V. B. Bajic and S. L. Klein, Brain, Behav., Immun., 2008, 22, 503–516 CrossRef CAS.
- S. L. Klein, S. Dhakal, R. L. Ursin, S. Deshpande, K. Sandberg and F. Mauvais-Jarvis, PLoS Pathog., 2020, 16, e1008570 CrossRef CAS.
- E. P. Scully, J. Haverfield, R. L. Ursin, C. Tannenbaum and S. L. Klein, Nat. Rev. Immunol., 2020, 20, 442–447 CrossRef CAS.
- L. Yang, S. Liu, J. Liu, Z. Zhang, X. Wan, B. Huang, Y. Chen and Y. Zhang, Signal Transduct. Target Ther., 2020, 5, 128 CrossRef CAS PubMed.
- T. Colombani, L. J. Eggermont, Z. J. Rogers, L. G. A. McKay, L. E. Avena, R. I. Johnson, N. Storm, A. Griffiths and S. A. Bencherif, Adv. Sci., 2021, 8, 2100316 CrossRef CAS PubMed.
- A. K. Blakney, Y. Jiang and K. A. Woodrow, Drug Delivery Transl. Res., 2017, 7, 796–804 CrossRef CAS.
- F. B. S. P. Duran, L. Burnett, S. A. Menefee, M. Cook, G. Zazueta-Damian, M. Dzieciatkowska, E. Do, S. French, M. M. Shah, C. Sanvictores, K. C. Hansen, M. Shtrahman, K. L. Christman and M. Alperin, 2021, bioRxiv DOI:10.1101/2021.05.28.446170.
- N. Tucker, J. J. Stanger, M. P. Staiger, H. Razzaq and K. Hofman, J. Eng. Fiber Fabr., 2012, 7, 63–73 CAS.
- R. Sharma, T. Garg, A. K. Goyal and G. Rath, Artif. Cells Nanomed. Biotechnol., 2016, 44, 524–531 CrossRef CAS PubMed.
- C. M. Mazure and D. P. Jones, BMC Womens Health, 2015, 15, 94 CrossRef PubMed.
- B. A. Aguado, J. C. Grim, A. M. Rosales, J. J. Watson-Capps and K. S. Anseth, Sci. Transl. Med., 2018, 10, eaam8645 CrossRef.
- J. F. Reckelhoff, Hypertension, 2001, 37, 1199–1208 CrossRef CAS PubMed.
- T. Andersson, A. Magnuson, I. L. Bryngelsson, O. Frobert, K. M. Henriksson, N. Edvardsson and D. Poci, Int. J. Cardiol., 2014, 177, 91–99 CrossRef PubMed.
- S. L. Klein, Parasite Immunol., 2004, 26, 247–264 CrossRef CAS PubMed.
- A. M. Unruh, Pain, 1996, 65, 123–167 CrossRef CAS.
- S. Y. So and T. C. Savidge, Front. Endocrinol., 2021, 12, 684096 CrossRef PubMed.
Footnote |
| † Equal contribution of authors. |
| This journal is © The Royal Society of Chemistry 2022 |