Multi-layered stimuli responsive DNA micelles for the stepwise controlled release of small molecules†
Joshua J.
Santiana
,
Shraddha S.
Sawant
 ,
Nicole
Gomez
and
Jessica L.
Rouge
,
Nicole
Gomez
and
Jessica L.
Rouge
 *
*
Department of Chemistry, University of Connecticut, Storrs, CT 06269, USA. E-mail: jessica.rouge@uconn.edu
First published on 28th February 2022
Abstract
Controllable release of multiple distinct cargoes from a nanomaterial is crucial to a variety of therapeutic and catalytic applications. In this study, we describe a DNA functionalized multi-layered surface crosslinked micelle (mlSCM) consisting of individually degradable layers. The DNA modified mlSCM has the ability to encapsulate separate small molecule cargo in distinct compartments within the nanocapsule, separated by chemical crosslinkers. Through a multistep self-assembly process, we show physical separation of internalized cargo as evidenced by electron microscopy, along with observation of chemical control over release, and chemical reaction conditions, as seen by fluorescence spectroscopy and a high-performance liquid chromatography mass spectrometry assay. Additionally, we evaluated the ability of these DNA crosslinked micelles to co-release two separate cargoes into the same cellular environment through an in vitro confocal microscopy assay. We show individualized targeting of two distinct but related dyes for the detection of ATP and mitochondria. The colocalization of these dyes indicates that unique locations and signals related to cellular respiration can be identified using a single mlSCM. Through these studies we ultimately show that the mlSCM has a tailorable design with the potential to be applied to numerous applications, ranging from sensing to drug delivery.
1. Introduction
Controlled release from a nanomaterial is a highly sought-after property. It can enable controlled reactivity of small molecules in solution and result in reactions and chemical signalling in solution that is unique from the bulk solution properties of molecules. Thus, this material property is helpful for a range of applications, from catalysis to sensing to drug delivery. Increasing the specificity of controlled release from nanoparticle systems can enable safer nanomedicines1,2 or limit chemical reactions at the nanoscale for enhanced sensing.3,4 In particular, developing customizable systems for combination therapies has become a central focus of nanoparticle formulation and design for pharmaceutical applications as efforts to deliver more than one drug to a single cell can enable synergistic therapeutic responses.3 In light of this, researchers have begun to develop systems that are compatible with combination therapies wherein two cargoes are incorporated within a single nanomaterial. Such nanomaterials include liposomes,4 polymer nanoparticle systems,5 dendrimers,6 and carbon nanotubes,7 among others. In many of these systems, multiple small molecule drugs are incorporated into the core of a material that can then be released from the nanomaterial by a single stimuli. This can lead to therapeutic responses that have improved pharmacological profiles, decreased toxicity, and better control over dosage ratios.3 Although significant progress has been made in these areas, the field still lacks a nanomaterial delivery system that can effectively control the separation of cargo during delivery and that can then sequentially release small molecules in response to specific stimuli.8 At the heart of such a design is the fundamental need for control over the nanoscale compartmentalization of cargo within the nanomaterial itself. Here, we introduce the design of a nanoscale system that is highly modular and can control the degradation, and therefore release, of individual cargo compartments within it. The experiments discussed within provide a proof-of-concept for the dual release of small molecule cargo from a single nanomaterial. This system is highly modular and can be tailored to release a variety of cargo from a single nanocapsule design.Our multi-layered nanomaterial was developed using a layer-by-layer self-assembly approach using surfactants, wherein the final product is soluble in aqueous media. The design takes advantage of our recently developed nanocapsule design that we refer to as a nucleic acid nanocapsule (NAN).9 The NAN is a micelle-based nanocapsule that can encapsulate hydrophobic compounds and can be crosslinked with a variety of synthetic crosslinkers by either copper(I)-catalyzed alkyne–azide cycloaddition (CuAAC) or UV-catalyzed thiol–yne click chemistry. The first iteration of the NAN was crosslinked with a diazido ester crosslinker and was shown to release cargo in the presence of the enzyme esterase.9 As esterase is a ubiquitous enzyme,10 we increased the NAN's specificity for proteolytic enzyme targets by introducing peptide crosslinkers.11 The crosslinkers that were selected are well-known enzyme substrates that can be cleaved under specific biochemical conditions.12,13 For example, we showed highly specific cargo release in the presence of matrix metalloproteinase 9 (MMP9), and a lack of release in the presence of other related proteases.11 With the ability to customize the crosslinker in the NAN system, it provides the option to design a nanocapsule that can be tailored to degrade under a variety of specific environmental conditions – a particularly useful design aspect when seeking individualized cargo release from a single nanomaterial.
When designing the mlSCM, the initial focus was to develop a strategy in which our alkyne-terminated surfactant would be allowed to form an additional layer around a NAN. Ultimately, we took inspiration from our previous work in which we encapsulated hydrophobically modified gold nanoparticles inside of an SCM.11 Ideally, if we could functionalize a NAN with a hydrophobically modified oligonucleotide ligand, we could utilize its hydrophobic nature to seed the encapsulation of the NAN within an additional layer of surfactant. To complete this, we investigated the use of a poly-T20 oligonucleotide tethered to cholesterol as a way to seed the assembly of a second layer within the assembled DNA micelle. The work discussed here focuses on the development and characterization of this mlSCM design as well as proving stimuli responsive cargo release from its individual compartments. Its design and synthesis is an important step towards being able to develop these complex systems in a robust and reproducible way.
2. Materials and methods
2.1. Materials
C12 surfactant, dansyl modified surfactant, and diazido ester crosslinker were synthesized in-house following previously reported protocols.9 All DNA was synthesized in-house on an ABI 394 automated DNA synthesizer using standard coupling procedures. Diazido PEG crosslinker (azide-PEG3-azide) was purchased from Lumiprobe and diazido disulfide crosslinker (azidoethyl-SS-ethylazide) was purchased from BroadPharm. 5-TAMRA (5-carboxytetramethylrhodamine) was purchased from Thermo Fisher. Fluorescein was purchased from Sigma-Aldrich. Cyanine 5 tetrazine was purchased Lumiprobe. TCO-PEG3-acid (trans-cyclooctene-polyethylene glycol-acid) was purchased from BroadPharm.2.2. Cholesterol modified nucleic acid nanocapsules (Chol-NANs)
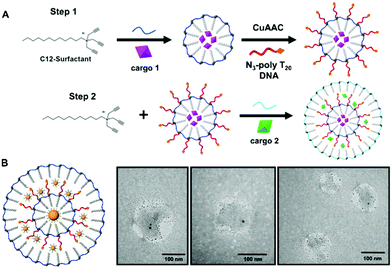
Surface crosslinked micelles (SCMs) were prepared as previously reported using the diazido ester crosslinker.9 All crosslinkers utilized in SCM syntheses can be found in Fig. S1 (ESI†). SCMs were diluted to a total volume of 500 μL to a concentration of 250 μM. Included in this dilution was SH-TTTTT-cholesterol (250 μM) and 2-hydroxy-4-(2-hydroxyethoxy)-2-methylpropiophenone (DHEMPP) (20 μM). The final reaction solution was placed in a Rayonet reactor and treated with UV light (365 nm) for 30 minutes. The product was purified by size exclusion chromatography and characterized by DLS and zeta potential. The stepwise assembly of the mlSCM and mlNAN is shown in Fig. 1A and Scheme S1 (ESI†), respectively.2.3. Multi-layered surface crosslinked micelles (mlSCMs)
Chol-NANs were lyophilized to remove remaining solvent. 0.9 mg (5 mM) of surfactant was added to the Eppendorf tube containing the chol-NANs. 455 μL of water and 0.25 mM of cargo (fluorescent dye) were added and the solution was mixed and sonicated for 30 minutes. A diazido crosslinker (6 mM) was dissolved in 5 μL DMSO and then added to the reaction mixture and was sonicated until fully dissolved. 10 μL (5 mM) of sodium ascorbate was added to the reaction mixture. A pre-mixed solution of copper(II) sulfate penta-hydrate (2.5 μL, 250 μM) and tris-hydroxypropyltriazolylmethylamine (THPTA) (5 μL, 500 μM) was added to the reaction mixture and left to stir at room temperature for 4 hours. The reaction was purified by NAP-5 sephadex column. 500 μL fractions were collected and analyzed by DLS and zeta potential measurements.2.4. Multi-layered nucleic acid nanocapsules (mlNANs)
mlSCMs were diluted to a total volume of 500 μL to a concentration of 250 μM. Included in this dilution was a thiolated DNA anchor (poly T20) (500 μM) and 2-hydroxy-4-(2-hydroxyethoxy)-2-methylpropiophenone (DHEMPP) (20 μM). The final reaction solution was placed in a Rayonet reactor and treated with UV light (365 nm) for 30 minutes. The product was purified by size exclusion chromatography and characterized by DLS and zeta potential.2.5. Fluorometric cargo release from SCMs and mlSCMs
A solution of ester crosslinked SCMs (5 μM, 5-TAMRA cargo) was prepared in water at a total volume of 800 μL. The sample was treated with sodium hydroxide and fluorescence measurements were taken. Further measurements were taken every 30 seconds. Excitation: 557 nm, emission: 583 nm. The same protocol was followed using mlSCMs (inner layer: 15 nm AuNP, outer layer: 5-TAMRA) to track the release of 5-TAMRA (5 μM mlSCMs). See Fig. 2A.2.6. Fluorometric cargo release from dansyl modified surfactant mlSCMs
A solution of ester crosslinked mlSCMs (inner layer: 15 nm AuNPs and fluorescein, outer layer: dansyl modified surfactant) (6.25 μM) was prepared in 10 mM Tris HCl buffer (pH 7.5) at a total volume of 800 μL. The solution was heated to 37 °C using an external Peltier unit. Esterase (Porcine liver esterase, Sigma Aldrich, 4 units) was added and an initial measurement was taken. Additional measurements were taken at the following timepoints: 5, 15, 30 and 60 minutes. In order to track breakdown and release of distinct layers, separate emission profiles were tracked. Dansyl release (excitation: 330 nm, emission scan: 360–625 nm). Fluorescein release (excitation: 488 nm, emission: 512 nm). See Fig. 2B.2.7. Fluorometric cargo co-release of 5-TAMRA and fluorescein
A solution of ester crosslinked mlSCMs (inner layer: 5-TAMRA, outer layer: fluorescein) (6.25 μM) was prepared in water at a total volume of 800 μL. The sample was treated with sodium hydroxide and an initial measurement was taken. Additional measurements were taken every 60 seconds for 35 minutes. In order to track the release of two different dyes from each layer, separate emission profiles were tracked. Fluorescein release (excitation: 488 nm, emission: 512 nm). 5-TAMRA release (excitation: 557 nm, emission: 583 nm). See Fig. S4 (ESI†).2.8. Fluorometric cargo release of ATP-Red and MitoTracker Green
A solution of ester crosslinked mlNANs (inner layer: ATP-Red, outer layer: MitoTracker Green) (5 μM) was prepared in 10 mM Tris HCl buffer (pH 7.5) with 5 μL ATP (2.5 mM) at a total volume of 200 μL. The solution was heated to 37 °C and esterase (Porcine liver esterase, Sigma Aldrich, 4 units) was added. An initial measurement was taken and additional measurements were taken every 30 seconds for 1 hour. In order to track breakdown and release of distinct layers, separate emission profiles were tracked. ATP-Red release (excitation: 510 nm, emission: 570 nm). MitoTracker Green release (excitation: 490 nm, emission: 516 nm). See Fig. S11 (ESI†).2.9. Fluorometric release of ATP-Red in the presence of ATP and GMP
A solution of ester crosslinked NANs containing ATP-Red (5 μM) was prepared in 10 mM Tris HCl buffer (pH 7.5) with 2.5 μL of either ATP (2.5 mM) or GMP (2.5 mM) at a total volume of 200 μL. The solution was heated to 37 °C and esterase (Porcine liver esterase, Sigma Aldrich, 4 units) was added. An initial measurement was taken and additional measurements were taken every 30 seconds for 1 hour. To track the release and the enhanced fluorescence of ATP-Red in the presence of ATP the following profile was used: excitation: 510 nm, emission: 570 nm). See Fig. S12 (ESI†).2.10. Tetrazine-TCO ligation protocol
10 μL (100 μM) of cyanine 5 tetrazine and 20 μL (200 μM) of TCO-PEG3-acid were added to 440 μL of water and were allowed to mix for one hour. The product was analyzed by HPLC and mass spectrometry. See Fig. S6 and S8 (ESI†).2.11. Reverse phase HPLC method for cyanine 5 tetrazine
During method development, a gradient was run at 1 mL min−1 from 0% to 100% water and 100% to 0% methanol over a 40 minute time period. The first sample injected was cyanine 5 tetrazine and the peak was observed at a retention time of about 10 minutes, or at 75% methanol, 25% water. Subsequent experiments were completed using isocratic elution at this solvent ratio over a period of eight minutes.2.12. Characterization of SCMs, NANs, mlSCMs and mlNANs
Dynamic light scattering and zeta potential measurements for nanocapsule characterization was completed using a Zetasizer Nano-ZS90. Transmission electron microscopy (TEM) images were taken using a FEI Tecnai 12 G2 Spirit Bio TWIN. Samples were stained with 0.5% uranyl acetate. Fluorometric analysis studies were completed using a Jobin Yvon Fluorolog 3 series fluorometer as well as a BioTek Synergy H1 microplate reader. HPLC studies were completed using a Waters 1525 system with a binary HPLC pump with a Waters 2475 multi wavelength fluorescence detector (excitation: 646 nm, emission: 662 nm). A C4 XBridge Protein BEH, 3.5 μM, 4.6 mm × 100 mm column was utilized.2.13. Confocal imaging of FITC mlNANs in A549 cells
A549 cells were cultured in F12K media supplemented with 10% FBS at 37 °C and 5% CO2. Confluent cells were plated in a ibidi μ-Slide 8 well chamber at a density of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and incubated overnight. A549 cells were then treated with 30 μM of mlNANs for 4 hours. After 4 hours, the cells were incubated with lysotracker green (75 nM) for 30 min followed by 5 min incubation with Hoechst stain. Lastly, all the wells were washed with 1× PBS and imaged using a Leica SP8 Confocal microscope. Fluorescence emission images were taken using laser lines of 405 nm, 488 nm and 561 nm to excite Hoechst 33342, FITC and Lysotracker respectively.
000 cells per mL and incubated overnight. A549 cells were then treated with 30 μM of mlNANs for 4 hours. After 4 hours, the cells were incubated with lysotracker green (75 nM) for 30 min followed by 5 min incubation with Hoechst stain. Lastly, all the wells were washed with 1× PBS and imaged using a Leica SP8 Confocal microscope. Fluorescence emission images were taken using laser lines of 405 nm, 488 nm and 561 nm to excite Hoechst 33342, FITC and Lysotracker respectively.
2.14. Confocal imaging of ATP-Red and MitoTracker Green loaded mlNANs in HeLa cells
HeLa cells were cultured in DMEM media supplemented with 10% FBS at 37 °C and 5% CO2. Once confluent, they were plated into Lab-Tek II 8 well confocal chambers at a density of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and incubated overnight. The cells were then treated 50 μM mlNANs, synthesized according to protocol 2.7 except made with ATP Red 1 in the interior and MitoTracker Green on the exterior, for 4 hours. Untreated and free dyes were used as controls. After incubation, the cells were washed with 1× PBS and imaged using a Leica SP8 Confocal microscope. Fluorescence emission images were taken using laser lines of 561 nm and 405 nm to excite Mito tracker/ATP Red 1 and Hoechst 33342, respectively.
000 cells per mL and incubated overnight. The cells were then treated 50 μM mlNANs, synthesized according to protocol 2.7 except made with ATP Red 1 in the interior and MitoTracker Green on the exterior, for 4 hours. Untreated and free dyes were used as controls. After incubation, the cells were washed with 1× PBS and imaged using a Leica SP8 Confocal microscope. Fluorescence emission images were taken using laser lines of 561 nm and 405 nm to excite Mito tracker/ATP Red 1 and Hoechst 33342, respectively.
3. Results and discussion
First, surface crosslinked micelles (SCMs) were synthesized using a diazido ester crosslinker as described previously.9 From there, thiol–yne click chemistry was utilized to functionalize the SCM surface with a cholesterol modified oligonucleotide, 5′-SH-TTTTT-cholesterol-3′ (SH-polyT5-chol). Once functionalized, the SH-polyT5-chol NANs are then characterized by dynamic light scattering (DLS) post size exclusion fractionation over a G-25 sephadex column (Fig. S2, ESI†). The solvent was removed via lyophilization and an additional layer of surfactant was introduced to the NANs at a concentration of 5 mM. The critical micelle concentration (CMC) of the surfactant is 10 mM, so decreasing the concentration allowed for the growth of an additional layer of surfactant around the hydrophobic NAN. From there, the mlSCMs were crosslinked with the diazido ester crosslinker via copper(I)-catalyzed azide alkyne cycloaddition (CuAAC) (Fig. 1A). The mlSCMs were purified via size exclusion chromatography and characterized via dynamic light scattering (DLS) and zeta potential measurements.3.1. TEM visualization of distinct mlSCM layers
Next it was of interest to track the encapsulation of cargo within each layer of the mlSCMs. A transmission electron microscopy (TEM) assay was developed using electron dense gold nanoparticles (AuNPs) so that the individual layers formed within the micelles could be easily visualized. Hydrophobically modified gold nanoparticles of differing sizes were selected (15 nm and 2 nm) to be encapsulated within specific compartments in order to visualize each layer. The synthetic protocol for encapsulating the AuNP within the micelle core of the NAN was adapted from our previously reported strategy.11 One initial concern with this strategy was the possibility of empty nanocapsule formation. However, using centrifugation one can easily separate the Au-mlSCMs from empty mlSCMs thanks to the difference in densities of the AuNPs encapsulated within the particles relative to empty particles. Centrifugation also serves to purify and remove any unreacted reagents from the click reaction and any unreacted crosslinker from the AuNP embedded SCMs. For the electron microscopy study, the inner layer of the mlSCMs encapsulated the 15 nm gold nanoparticles and was crosslinked with a diazido PEG crosslinker. This SCM was then functionalized with SH-polyT5-cholesterol and brought forward for encapsulation with an additional layer of surfactant in the presence of a smaller 2 nm gold nanoparticles. The outer layer of the micelle was also crosslinked with the diazido PEG crosslinker. The final purified mlSCMs were brought forward for TEM analysis via staining with uranyl acetate on a carbon mesh grid. As can be seen in Fig. 1B and Fig. S3 (ESI†), the gold nanoparticles can be observed in distinct locations within the mlSCM construct. While individual layers cannot be directly observed by TEM, the location of the gold nanoparticles provided us with structural information about this system, indicating two distinct layers. The larger gold nanoparticles were found centrally in the majority of the mlSCMs while the smaller gold nanoparticles were found spread out around the perimeter. This indicated that we were able to successfully encapsulate multiple cargoes within separate compartments of each nanocapsule.While this was an important step in the validation and characterization of the mlSCMs, it was of interest to analyze whether we could induce stimuli responsive cargo release from each individual layer of the mlSCMs. To complete this analysis and confirm control over cargo release, a number of assays were developed, including fluorescence and high-performance liquid chromatography (HPLC) based studies.
3.2. Dye release from individual layers of a mlSCM
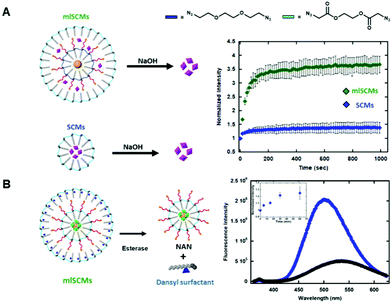
The first assay investigated the release profiles of a dye from a multi-layered SCM as compared to that of a SCM. To enhance any fluorescence signal differences between the two constructs we incorporated an AuNP cargo within the inner layer of the mlSCM. AuNPs can quench fluorescence of various dyes when in close proximity to one another.14,15 This effect is due to the localized surface plasmon resonance (SPR) exhibited by AuNPs that can interfere with fluorescence pathways of nearby molecules.16 Specifically, it has been shown that a number of fluorescent dyes can be significantly quenched in the presence of gold nanoparticles at up to a distance of 30 nm.17 As our micelle compartments appear to be separated by less than 30 nm based on TEM, we hypothesized that incorporating an AuNP into the inner compartment would quench the dye signal in the mlSCM's outer layer until it is released, thereby providing a larger dynamic range in signal turn on for our assay upon the nanocapsules degradation.To this end, a hydrophobically modified gold nanoparticle (dodecylamine modified AuNP) was encapsulated within the inner layer of the mlSCM in order to quench the fluorescence of a neighbouring dye molecule (5-TAMRA), encapsulated within the particles outer layer. From here, we expected to see an enhanced release profile of 5-TAMRA from the outer layer of the mlSCM when exposed to the appropriate stimulus in comparison to 5-TAMRA release from a standard SCM without AuNPs. The mlSCM construct synthesized for this assay encapsulated 15 nm hydrophobically modified gold nanoparticles in the inner layer and was crosslinked with a diazido PEG crosslinker preventing its release. In contrast, 5-TAMRA was encapsulated in the outer layer and crosslinked with a degradable diazido ester crosslinker. A fluorometric assay was then completed (excitation: 557 nm, emission: 583 nm) to track the release of 5-TAMRA from each construct in the presence of 1 M NaOH. The results of this fluorometric assay (Fig. 2A) indicated that we do in fact observe an enhanced release profile of 5-TAMRA from the mlSCMs due to the proximity of the AuNP in the inner core and thus its initial quenched state. Each of the constructs were analyzed at an initial concentration of 5 μM mlSCM (meaning 125 nM 5-TAMRA due to 2.5% loading). The results show a more significant fluorescence intensity increase of 5-TAMRA released from the mlSCMs as compared to the release from the SCMs alone. Such a result is a useful way to enhance the signal of dye release from a micelle for sensing applications. Ultimately, this assay indicated that there are in fact two layers of surfactant within the mlSCM construct, and that each can contain separate cargo. It was also shown that the cargo of one layer can influence structural and optical properties of other cargo within the construct.
3.3. Individual stepwise release of molecular cargo from each mlSCM layer
Our next goal was to investigate our ability to control the breakdown and release of cargo from each individual layer of the mlSCMs.In order to test this, we utilized a dye labeled surfactant synthesized by our group for tracking micelle assembly states.18 This new surfactant utilizes a dansyl group as a chemical probe. Dansyl is a useful probe for micelle assembly as it is environmentally sensitive; in aqueous environments it fluoresces weakly (max em. 510 nm), while in nonpolar environments it fluoresces strongly and a blueshift is observed (max em. 540 nm). Thus, through the fluorometric analysis of this surfactant, it can be determined whether or not micelles are present in a solution of surfactants. Using the dansyl modified surfactant, a fluorescence-based assay was developed for tracking both the assembly and breakdown of individual mlSCM compartments.
For the construct synthesized for these studies, the inner layer encapsulated both hydrophobic gold nanoparticles and fluorescein (to quench the fluorescein initially) and its surface was crosslinked with a diazido ester crosslinker. The outer layer was then assembled using the dansyl modified surfactant and contained no cargo. This outer layer was also crosslinked with the diazido ester crosslinker (Fig. 2B). In order to track both micelle degradation and cargo release from each layer, the mlSCMs were treated with esterase (4 units) and the solution's fluorescence tracked over time. The dansyl emission (excitation: 330 nm, emission: 360–625 nm) was monitored as a way to track the intact or degraded state of the outer layer's micelle, and cargo release from the inner layer was monitored through tracking release of fluorescein (excitation: 488 nm, emission: 508–625 nm) from the micelles inner core. Since both layers are crosslinked with the diazido ester crosslinkers, they should both degrade in the presence of esterase. We anticipated observing two separate changes in the fluorescence profile of this system to indicate successful cargo release from each compartment over time. As anticipated, the degradation of the outer layer was observed as indicated by a blueshift from 510 nm to 540 nm as well as a decrease in the overall fluorescence intensity as seen in Fig. 2B. The degradation of the inner layer was also observed by the observation of an increase in the fluorescence intensity of fluorescein over time (inset Fig. 2B). This assay confirmed the formation of the outer layer of the mlSCM and its ability to be degraded by an external stimulus, followed by the subsequent release of the inner layer's cargo.
Having established the multi-layered nature of the mlSCMs by a combination of TEM and fluorescence assays, we set out to observe the release of small molecule cargo from the individual layers of the nanocapsule. To do this, a mlSCM nanocapsule was assembled using the alkyne-terminated surfactant and was degraded by hydrolysis with NaOH. Here, the diazido ester crosslinker was used to crosslink both the inner and outer layers. 5-TAMRA was encapsulated in the inner layer while fluorescein was encapsulated in the outer layer. As the breakdown of the mlSCM quickly occurs in the presence of NaOH, rapid measurements were taken to track the amount of time that it took for all cargo to be released from each individual compartment based on the emission wavelength of the cargo. Fluorometric analysis was completed after treatment of mlSCMs (6.25 μM) with 500 μM NaOH. The change in fluorescence intensity of fluorescein (excitation: 488 nm, emission: 508 nm) and 5-TAMRA (excitation: 557 nm, emission: 583 nm) was then monitored over time. The results of this assay showed that there is indeed a difference in the time it took for total release of cargo from each layer. It took about 2 minutes for complete release of fluorescein from the outer layer while it took about 6 minutes for complete release of 5-TAMRA from the inner layer (Fig. S4, ESI†). This assay indicated the that the release of the cargo from each layer of the mlSCM may occur in a subsequent fashion rather than all at once and may ultimately be tuned based on the chemical nature of the crosslinker. As the crosslinker here was the same, we interpret the lag between total release from both layers to be due to delayed physical access of the inner layer when treated with base. We therefore anticipate that with two different crosslinkers the timing of each layer's degradation can be fine-tuned. Based on this result we next set out to see if we could use our mlSCM to control the reactivity of two substrates via their encapsulation within separate compartments of the mlSCMs.
3.4. Controlling the reaction of two small molecules encapsulated within the mlSCM
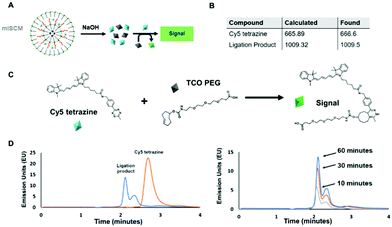
Using two molecules that are known to react together, we sought to understand the effects their physical separation at the nanoscale would have on their relative reactivity with each other.To carry out this assay, we utilized an HPLC in order to monitor the reaction between the two small molecules after their release. We chose molecules that were hydrophobic enough to be encapsulated within a micelle, but could also be released into aqueous solution and readily react with one another. In addition, the compounds needed to by optically active so that we could easily track them via UV and fluorescence monitors. To meet these criteria, we chose two reactants that undergo a trans-cyclooctene (TCO) – tetrazine ligation (Fig. 3A and C and Fig. S5, ESI†). This technique is based on an inverse-demand Diels–Alder cycloaddition between TCO and tetrazine, which forms a dihydropyridazine bond.19 This bioconjugation strategy occurs under physiological conditions and has ultrafast kinetics (up to 105 M−1 s−1) making it a suitable proof-of-concept system for assessing the reactivity of cargo within individual layers of the mlSCMs.20 More specifically, cyanine 5 tetrazine was selected as the first reactant and TCO-PEG3-acid as the second, which, post ligation would allow for a change in the retention time observed via HPLC after successful product formation. We identified a diazido disulfide crosslinker as an appropriate stimuli responsive crosslinker to incorporate into our mlSCM construct for this assay as it can be readily degraded using dithiolthreitol (DTT) and would not affect the reactivity of the substrates in our reaction.21,22
First, control studies were completed to track starting materials and product formation by mass spectrometry (MS) and reverse phase HPLC. The fluorescence intensity of the cyanine 5 was tracked using a fluorescence detector (excitation: 646 nm, emission: 662 nm). The retention times were determined to be 3.6 minutes for cyanine 5 tetrazine and 2.1 minutes for the ligation product (Fig. 3D), and product formation was confirmed by MS (Fig. S8, ESI†). From here, it was important to track the formation of the product after release from individual mlSCM compartments. The first mlSCM batch that was assembled for these studies contained TCO-PEG3-acid in the inner layer and cyanine 5 tetrazine in the outer layer, while both layers were crosslinked with the diazido disulfide crosslinker. The construct was treated with DTT (500 μM) and analyzed by HPLC and MS after 6 hours (Fig. 3B and D). The ligation product was confirmed to be released from the mlNANs by MS (Fig. S9, ESI†) and its retention time was 2.1 minutes, collectively indicating successful release and reaction of both cargoes. Next, a time course analysis was conducted. An initial run (prior to DTT addition) was completed as a control and after addition of DTT, HPLC analysis was completed every 10 minutes. As the TCO-tetrazine ligation is very rapid, most of the product formation occurs within the first 10 minutes, as can be observed in Fig. 3D. Ultimately, this assay showed successful cargo release and product formation over time.
For the next HPLC study, we wanted to prove successful control over release of cargo from specific compartments within the mlSCM. This mlSCM batch was assembled to contain TCO-PEG3-acid in the inner layer (diazido PEG crosslinker) and cyanine 5 tetrazine in the outer layer (diazido disulfide crosslinker). Through this analysis, we expected to observe release of cyanine 5 tetrazine from the outer layer in the presence of DTT, but a lack of release of TCO-PEG3-acid from the inner layer. Thus, leading to a lack of formation of the ligation product. The results show that the cyanine 5 tetrazine remains consistent over the course of an hour (Fig. S7 and S10, ESI†). This indicated release of cyanine 5 tetrazine with a lack of reactivity with TCO-PEG3-acid. Overall, these studies showed that we have control over the release of cargo from each mlSCM compartment at the nanoscale.
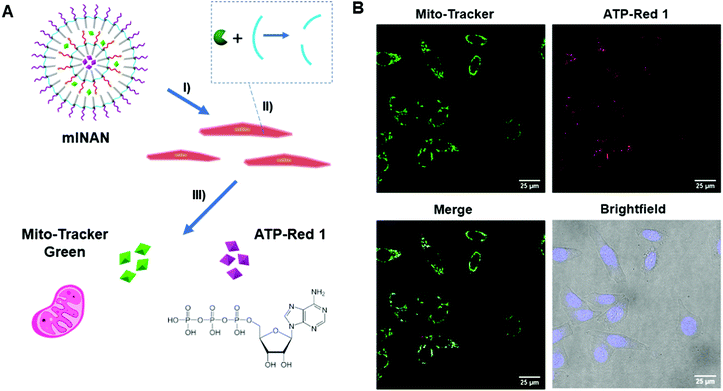
3.5. Dual cellular probe delivery using a multi-layered nucleic acid nanocapsule (mlNAN)
Having shown control over the degradation of the individual layers of the mlSCMs, we ultimately wanted to investigate its utility in a biological system. Having the ability to deliver drugs and dyes in combinations to cells has great value for both fundamental and applied biomedical research. As a basic proof-of-concept we incorporated two different dyes, ATP-Red23 and Mito Tracker Green within the individual layers of our particle, further functionalized it with DNA, and converted it into a multi layered nucleic acid nanocapsule (mlNAN) in order to enhance its cellular uptake. ATP-Red is a recently developed dye that is highly specific for ATP within the cell. This dye was selected as we anticipated that it could be readily encapsulated within our micelles and it would provide a functionality to our system by measuring intracellular ATP levels. Changes in fluorescence intensity (based on ATP concentration) can be observed by confocal microscopy. For example, with the introduction of camptothecin, a cytotoxic drug, intracellular ATP concentrations are increased due to apoptosis.24 The second dye, MitoTracker Green, was selected as it localizes to the mitochondria, where ATP is produced. Also due to their differences in emission wavelength, ATP-Red and MitoTracker Green can be observed separately and analyzed for their degree of colocalization. Prior to incubating these multi-dye loaded NANs in cells, we first incubated an empty mlNAN functionalized with a FITC-labeled polyT20 DNA in cells (A549) as a control to assess their degree of internalization and it was noted that mlNANs underwent endocytosis similar to single layered NANs as evidenced by the DNA's FITC signal colocalizing with lysotracker red staining of the endosomes (Fig. S13, ESI†).Next we investigated the localization of the multi-dye loaded mlNANs. For this assay, a double layered system was assembled in which ATP-Red was encapsulated within the inner layer while MitoTracker Green was encapsulated within the outer layer of an mlNAN construct. Both layers were crosslinked with the diazido-ester crosslinker. Initial fluorescence measurements indicated the successful incorporation and release of ATP-Red and MitoTracker Green (Fig. S11, ESI†) and that ATP-Red was specific for ATP (Fig. S12, ESI†). HeLa cells were incubated with these mlNANs (50 μM) for 4 h to allow for cellular uptake. They were then brought forward for analysis by confocal microscopy.
As can be observed in Fig. 4, fluorescence from both ATP-Red and MitoTracker Green was found within the HeLa cells mitochondria. This can be observed in individual channels as well as within the overlay merged images. The high degree of colocalization is in line with the expectation that ATP would be in the vicinity of the mitochondria, and indicates that there was successful release of two different cargoes from the mlNAN under intracellular conditions. With the completion of this analysis, we were able to confirm successful control over release and delivery of small molecules from individual layers of the mlNAN construct to the cellular space. This further proved the utility of the mlNAN as a delivery vehicle and opens the possibility of drug delivery as a future application of this construct.
4. Conclusions
In summary, we have successfully developed a multi-layered nanocapsule that has the ability to encapsulate and release multiple cargoes in a stimuli responsive manner. Through these proof-of-concept studies, we have shown that we have excellent control over the release of individual cargo from distinct compartments and the ability to control the release of small molecules for both chemical reactions and for delivery to a cellular environment. This construct has potential for applications such as therapeutics, diagnostics and biosensing. As a therapeutic, it can be utilized for combination therapy in the delivery of different small molecule drugs to separate cellular locations. For example, anti-inflammatory drugs can be delivered outside of the cell while anti-cancer drugs can be delivered to the cytosol. We have previously shown synergistic delivery of siRNA and small molecule drugs,25 which can also be implemented within this multi-layered nanocapsule. This construct has the potential to be highly modular in that a number of unique small molecules, crosslinkers and therapeutic oligonucleotides can be incorporated in a mix and match fashion.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Funding sources
The authors are grateful to funding from the NSF Grant 1847869 for supporting this work.Abbreviations
| mlSCMs | Multi layered surface crosslinked micelles |
| mlNANs | Multi layered nucleic acid nanocapsules |
| TEM | Transmission electron microscopy |
| SEM | Scanning electron microscopy |
| DLS | Dynamic light scattering |
| HPLC | High performance liquid chromatography |
| DTT | Dithiothreitol |
| TCO | trans-Cyclooctene |
| 5-TAMRA | 5-Carboxytetramethylrhodamine |
| NAN | Nucleic acid nanocapsule |
| CuAAC | Copper(I)-catalyzed azide–alkyne cycloaddition |
| DNA | Deoxyribonucleic acid |
| siRNA | Small interfering ribonucleic acid |
| CMC | Critical micelle concentration. |
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors would also like to thank the Zhao group at the University of Connecticut for providing dodecyl modified AuNPs for these studies.References
- N. Kamaly, B. Yameen, J. Wu and O. C. Farokhzad, Chem. Rev., 2016, 116, 2602–2663 CrossRef CAS PubMed.
- F. Meng, R. Cheng, C. Deng and Z. Zhong, Mater. Today, 2012, 15, 436–442 CrossRef CAS.
- S. Gadde, Med. Chem. Commun., 2015, 6, 1916–1929 RSC.
- J. Meng, F. Guo, H. Xu, W. Liang, C. Wang and X.-D. Yang, Sci. Rep., 2016, 6, 22390 CrossRef CAS PubMed.
- R. Yang, G. Mondal, D. Wen and R. I. Mahato, Nanomedicine, 2017, 13, 391–401 CrossRef CAS PubMed.
- J. Pan, L. P. Mendes, M. Yao, N. Filipczak, S. Garai, G. A. Thakur, C. Sarisozen and V. P. Torchilin, Eur. J. Pharm. Biopharm., 2019, 136, 18–28 CrossRef CAS PubMed.
- G. Biagiotti, F. Pisaneschi, S. T. Gammon, F. Machetti, M. C. Ligi, G. Giambastiani, G. Tuci, E. Powell, H. Piwnica-Worms, E. Pranzini, P. Paoli, S. Cicchi and D. Piwnica-Worms, J. Mater. Chem. B, 2019, 7, 2678–2687 RSC.
- J. K. Patra, G. Das, L. F. Fraceto, E. V. R. Campos, M. P. Rodriguez-Torres, L. S. Acosta-Torres, L. A. Diaz-Torres, R. Grillo, M. K. Swamy, S. Sharma, S. Habtemariam and H. S. Shin, J. Nanobiotechnol., 2018, 16, 71 CrossRef PubMed.
- J. K. Awino, S. Gudipati, A. K. Hartmann, J. J. Santiana, D. F. Carins-Gibson, N. Gomez and J. L. Rouge, J. Am. Chem. Soc., 2017, 139, 6278–6281 CrossRef CAS PubMed.
- D. Gilham and R. Lehner, Methods, 2005, 36, 139–147 CrossRef CAS PubMed.
- J. J. Santiana, B. Sui, N. Gomez and J. L. Rouge, Bioconjugate Chem., 2017, 28, 2910–2914 CrossRef CAS PubMed.
- A. Page-McCaw, A. J. Ewald and Z. Werb, Nat. Rev. Mol. Cell Biol., 2007, 8, 221–233 CrossRef CAS PubMed.
- V. Turk, V. Stoka, O. Vasiljeva, M. Renko, T. Sun, B. Turk and D. Turk, Biochim. Biophys. Acta, Proteins Proteomics, 2012, 1824, 68–88 CrossRef CAS PubMed.
- E. Dulkeith, A. C. Morteani, T. Niedereichholz, T. A. Klar, J. Feldmann, S. A. Levi, F. C. J. M. van Veggel, D. N. Reinhoudt, M. Moller and D. I. Gittins, Phys. Rev. Lett., 2002, 89, 203002 CrossRef CAS PubMed.
- S. Li, T. Zhang, Z. Zhu, N. Gao and Q.-H. Xu, RSC Adv., 2016, 6, 58566–58572 RSC.
- X. Huang and M. A. El-Sayed, J. Adv. Res., 2010, 1, 13–28 CrossRef.
- P. Reineck, D. Gomez, S. H. Ng, M. Karg, T. Bell, P. Mulvaney and U. Bach, ACS Nano, 2013, 8, 6636–6648 CrossRef PubMed.
- M. Arifuzzaman, A. K. Hartmann and J. L. Rouge, RSC Adv., 2020, 10, 42349–42353 RSC.
- K. Johann, D. Svatunek, C. Seidl, S. Rizzelli, T. A. Braun, K. Koynov, H. Mikula and M. Barz, Polym. Chem., 2020, 11, 4396–4407 RSC.
- K. Lang and J. W. Chin, ACS Chem. Biol., 2014, 9, 16–20 CrossRef CAS PubMed.
- H. I. Chiu, A. D. Ayub, S. N. A. M. Yusuf, N. Yahaya, E. A. Kadir and V. Lim, Pharmaceutics, 2020, 12, 38 CrossRef CAS PubMed.
- T. Fuoco, D. Pappalardo and A. Finne-Wistrand, Macromolecules, 2017, 50, 7052–7061 CrossRef CAS.
- L. Wang, L. Yuan, X. Zeng, J. Peng, Y. Ni, J. C. Er, W. Xu, B. K. Agrawalla, D. Su, B. Kim and Y.-T. Chang, Angew. Chem., Int. Ed., 2016, 55, 1773–1776 CrossRef CAS PubMed.
- M. V. Zamaraeva, R. Z. Sabirov, E. Maeno, Y. Ando-Akatsuka, S. V. Bessonova and Y. Okada, Cell Death Differ., 2005, 12, 1390–1397 CrossRef CAS PubMed.
- A. K. Hartmann, S. Gudipati, A. Pettenuzzo, L. Ronconi and J. L. Rouge, Bioconjugate Chem., 2020, 31, 1063–1069 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: All DNA sequences, small molecule crosslinkers, additional nanomaterial characterization and assembly protocols, fluorescence assay data, and buffer information can be found in the supporting information. See DOI: 10.1039/d1tb02722k |
| This journal is © The Royal Society of Chemistry 2022 |