Stimuli-responsive nucleic acid nanostructures for efficient drug delivery
Changping
Yang†
ac,
Xiaohui
Wu†
ab,
Jianbing
Liu
 *ab and
Baoquan
Ding
*ab and
Baoquan
Ding
 *abc
*abc
aCAS Key Laboratory of Nanosystem and Hierarchical Fabrication, National Center for NanoScience and Technology, 11 BeiYiTiao, ZhongGuanCun, Beijing 100190, China. E-mail: dingbq@nanoctr.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cSchool of Materials Science and Engineering, Henan Institute of Advanced Technology, Zhengzhou University, Zhengzhou 450001, China
First published on 25th November 2022
Abstract
Based on complementary base pairing, nucleic acid molecules have acted as engineerable building blocks to prepare versatile nanostructures with unique shapes and sizes. Benefiting from excellent programmability and biocompatibility, rationally designed nucleic acid nanostructures have been widely employed in biomedical applications. With the development of the chemical biology of nucleic acids, various stimuli-responsive nucleic acid nanostructures have been constructed by tailored chemical modification with multifunctional components. In this minireview, we summarize the representative and latest research about the employment of stimuli-responsive nucleic acid nanostructures for drug delivery in response to endogenous and exogenous stimuli (redox gradient, pH, nuclease, biomacromolecule, and light). We also discuss the broad prospects and remaining challenges of nucleic acid nanotechnology in biomedical applications.
Introduction
Stimuli-responsive drug delivery systems (DDSs) have been developed for the efficient delivery of small molecules and biomacromolecular drugs to target sites in space–time- and dosage-controlled manners in the past decades.1,2 Both endogenous and exogenous stimuli have been utilized to construct the stimuli-responsive DDSs.3–5 The main goals of ideal stimuli-mediated DDSs include good biocompatibility, low side effects, retention of cargo activity during the upload and delivery processes, desired cellular uptake efficiency, and controllable release of payloads at target sites.6 To prepare various kinds of stimuli-triggered DDSs, various carriers with diverse sizes and surface properties have been developed, including liposomes, cationic polymers, and inorganic particles.7–10 However, these carriers show some inadequate biocompatibility and certain unavoidable inherent cytotoxicity, mainly resulting from their composition, which is extrinsic to cells. Therefore, constructing stimuli-responsive DDSs with splendid biocompatibility is needed.As natural biomacromolecules, nucleic acids, including deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), play essential roles in many vital biological functions by storing and transferring genetic information.11 In 1982, Seeman first proposed a constructing strategy for nucleic acid nanostructures using DNA as building blocks and presented its potential to prepare ordered architectures and crystal lattices (Fig. 1a).12,13 This pioneering work has been recognized as the origin of DNA nanotechnology. In 2005, Turberfield and co-workers reported a mechanically robust DNA tetrahedron with a well-defined structure self-assembled by four oligonucleotides complementary to each other (Fig. 1b).14 Later, in 2006, Rothemund invented the DNA origami technique by folding a long single-stranded DNA (ssDNA, scaffold strand) with over 200 short ssDNAs (staple strands) to produce predesigned nanoscale patterns, including squares, disks, five-pointed stars, and so on (Fig. 1c).15 Apart from the unmodified nucleic acid molecules, branched DNA monomers were synthesized by connecting multiple DNA strands to chemical linkers. Branched DNA with different functional groups was then used to construct three-dimensional (3D) DNA nanostructures (Fig. 1d).16–20 Furthermore, rolling circle amplification (RCA) or rolling circle transcription (RCT) has paved an avenue for the production of hydrogels, nanoflowers, and nanosponges, which have been developed as drug carriers with high loading capacities (Fig. 1e).21–26 Some heterogeneous systems consisting of nucleic acids and inorganic nanoparticles have been prepared as well. Spherical nucleic acid (SNA), first introduced by Mirkin, is a typical representative constructed by co-assembly between the nucleic acids and inorganic nanoparticles (Fig. 1f).27,28 Encouraged by the previous developments in nucleic acid self-assembly, the assembled biocompatible nucleic acid nanostructures have been widely employed in biomedical applications.
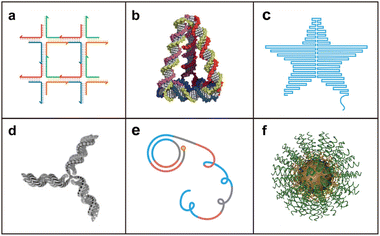 | ||
| Fig. 1 Representative strategies for the assembly of nucleic acid nanostructures. (a) DNA tile. Reproduced with permission.12 Copyright 1982, Elsevier. (b) DNA tetrahedron. Reproduced with permission.14 Copyright 2005, American Association for the Advancement of Science. (c) DNA origami. Reproduced with permission.15 Copyright 2006, Nature Publishing Group. (d) Branched DNA. Reproduced with permission.16 Copyright 2002, Nature Publishing Group. (e) DNA nanostructures based on rolling circle amplification. Reproduced with permission.21 Copyright 2012, Nature Publishing Group. (f) Spherical nucleic acid. Reproduced with permission.28 Copyright 2012, American Chemical Society. | ||
The nature of Watson–Crick base pairing endows nucleic acid nanostructures with the merits of flexibility and programmability of design and also spatial addressability.29 Therefore, the modularity of nucleic acid nanostructures is the maximal distinction towards other biomaterials. To date, various precisely designed 2D/3D nucleic acid nanostructures have been extensively constructed for biomedical applications involving intercellular communication, bioimaging, biosensing, and drug delivery.6,30–34 In addition, nucleic acid nanostructures can be easily modified with various stimuli-responsive elements, such as disulfide linkages, DNA triplex, i-motif, targeting aptamers, photo-cleavable bonds, and so on.35–43 Based on the advantages of good biocompatibility, structural programmability, spatial addressability, and convenient chemical modifiability, versatile nucleic acid nanostructures have been widely developed as stimuli-responsive drug delivery systems.
Redox gradient-responsive strategy
Disulfide linkage (–S–S–) is cleavable in the presence of reductants, such as glutathione (GSH), an intracellular reducing molecule.44–46 The concentration of the reducing agent (GSH) is different in blood plasma (2–20 μM) and cytosol (∼10 mM) and is even higher in cancer cells.47,48 Based on the concentration difference of GSH, nucleic acid nanostructures containing disulfide linkages can be relatively stable in the extracellular environment and cleaved by intracellular GSH to achieve a stimuli-responsive drug release in the cytosol.Some groups have reported several carrier-free strategies by introducing disulfide linkages in nucleic acid templates to form nanostructures with further enhanced cellular uptake and GSH-mediated cargo release. In 2015, Tan and co-workers proposed size-controllable and redox gradient-responsive DNA nanohydrogels co-assembled by two Y-shaped monomers and a DNA linker with sticky ends (Fig. 2a).49 The disulfide linkage modification in the three building units endowed nanohydrogel vectors with stimuli-responsive properties. In the presence of intracellular GSH, the nanohydrogels decomposed into antisense against c-raf-1 mRNA and DNAzymes targeting matrix metalloproteinase-9 (MMP-9) to efficiently inhibit cell proliferation and migration in A549 cells. Later, Yang and co-workers developed endogenous GSH-responsive oligonucleotide nanospheres by organizing guanidinium-containing disulfide monomers on the DNA templates via multiple salt bridges (Fig. 2b).50 The nanospheres can be directly delivered and accumulated in the cytosol by a thiol-mediated cellular uptake mechanism and further depolymerized under a reductive environment to release the oligonucleotides (antisense) for efficient gene silencing. These reports demonstrated that modification of disulfide linkage is an efficient strategy for redox gradient-responsive drug release.
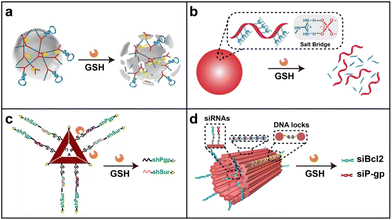 | ||
| Fig. 2 Redox gradient-responsive strategy. (a) A DNA nanogel for stimuli-responsive antisense delivery. Reproduced with permission.49 Copyright 2015, American Chemical Society. (b) An oligonucleotide nanosphere for antisense delivery. Reproduced with permission.50 Copyright 2019, Wiley-VCH GmbH. (c) A triangular DNA origami for efficient delivery of shRNA templates and the chemo-drug doxorubicin (DOX). Reproduced with permission.52 Copyright 2018, Wiley-VCH GmbH. (d) A tubular DNA nanodevice for siRNA and DOX co-delivery. Reproduced with permission.53 Copyright 2021, Wiley-VCH GmbH. | ||
Besides the co-assembled DNA complexes, the disulfide linkage can also be utilized in structurally well-defined DNA nanostructures. In 2018, Ding and co-workers constructed a functionalized double-bundle DNA tetrahedron structure containing a disulfide linkage and a nuclear localization signal (NLS) peptide to load antisense oligonucleotides (ASOs) for targeted and efficient antisense delivery.51 The rigidity and stability of the double-bundle DNA tetrahedron improved the enzyme resistance of the loaded ASOs. In response to an intracellular reducing environment, the antisense strands were released from the tetrahedron in the nucleus and cytoplasm for the downregulation of the target proto-oncogene c-raf. DNA origami involving abundant programmable sites is also an ideal candidate to construct stimuli-responsive DDSs. In 2018, Ding and co-workers used doxorubicin (DOX)-loaded triangular DNA origami to precisely organize two small hairpin RNA (shRNA) transcription templates targeting two functional genes (P-glycoprotein and survivin) for synergistic RNAi-/chemotherapy of multidrug-resistant (MDR) tumors (Fig. 2c).52 Incorporated with a cell-targeting aptamer and GSH-triggered disulfide linkage, this co-delivery nanoplatform efficiently inhibited MDR-tumor growth without obvious side effects.
Recently, they precisely encapsulated two types of small interfering RNAs (siRNAs) within the inner cavity of a DOX-loaded DNA nanodevice for combined cancer therapy (Fig. 2d).53 Stimulated by intracellular GSH, the nanodevice containing the disulfide linkage achieved mechanical opening and further the controllable release of siRNAs for localized gene silencing at the tumor site. With the modification of tumor-penetrating ligands, this DNA nanodevice demonstrated potent antitumor activity without observable systematic toxicity. These research studies provided a promising strategy for combined tumor therapy in vitro and in vivo by rationally employing stimuli-responsive elements.
pH-responsive strategy
In comparison with healthy tissues (pH ∼7.4), the tumor microenvironment (pH ∼6.5) is more acidic.8,54,55 Meanwhile, most drug carriers are internalized into cells through the endocytosis pathway. The pH values of sub-cellular compartments (endosome and lysosome) are lower than that of the extracellular environment and can even reach a level of 5.0.56,57 This difference in pH values is a main inspiration to design drug delivery systems in response to pH. Most pH-responsive delivery strategies of nucleic acid nanostructures have been designed by employing pH-sensitive sequences (DNA triplex and i-motif).In 2004, a pH-triggered DNA nanomachine based on a DNA triplex was described by Mao and co-workers.58 Similarly, Zhu and co-workers developed a pH-triggered triplex-DNA nanoswitch by immobilizing anti-MUC1 aptamers, small molecule drugs (DOX and cisplatin), and antisense DNA targeting survivin mRNA on the surface of Au nanoparticles for synergistic chemo- and gene-therapy (Fig. 3a).59 This nanoswitch kept a linear conformation under physiological pH conditions and changed to a triplex structure under an acidic environment. After the delivery process, the loaded drugs were controllably released at the tumor sites. Recently, Ding and co-workers constructed a tubular DNA nanodevice to precisely load an antigen peptide and two molecular adjuvants towards the toll-like receptor (CpG motifs and dsRNA) into its inner cavity locked by the pH-activated DNA strands (Fig. 3b).60 After entering the immune cells, the DNA locks of this nanodevice transitioned from a duplex into a triplex structure under an endosomal acidic microenvironment, resulting in the release of the antigen and two adjuvants to elicit a potent immune response and long-term protection against tumor rechallenge. This study demonstrated the great potential for producing personalized cancer vaccines based on the stimuli-responsive DNA origami technique.
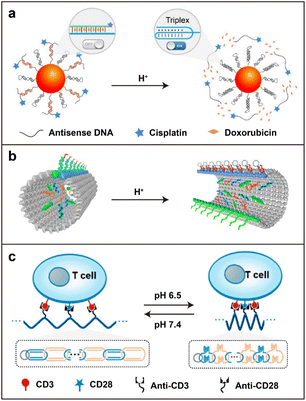 | ||
| Fig. 3 The pH-responsive strategy. (a) A triplex DNA nanoswitch for a pH-sensitive release of multiple cancer drugs. Reproduced with permission.59 Copyright 2019, American Chemical Society. (b) A tubular DNA origami locked by pH-triggered DNA strands for the co-assembly of antigens and adjuvants inside. Reproduced with permission.60 Copyright 2021, Nature Publishing Group. (c) A pH-driven interlocked DNA nano-spring to stimulate T-cell activation in vivo. Reproduced with permission.62 Copyright 2022, American Chemical Society. | ||
Besides DNA triplex, the pH-responsive DNA i-motif is also developed for biomedical applications. In 2013, Balasubramanian and co-workers reported a pH-driven DNA nanostructure based on the DNA i-motif.61 When the pH changes from alkaline to acidic, the protonated cytosine (C) residues in the stretched i-motif (open state) are incorporated with unprotonated C to form base pairs and further interdigitated with each other to construct the stable quadruple helix (close state).61 This conformational change in nucleic acid sequences driven by pH changes attracted lots of interest with regard to biosensing and controllable drug delivery.57,63 Specific activation of T-cell proliferation in tumors is crucial for reducing nonspecific inflammation and autoimmunity in antitumor immunotherapy.64 To address this issue, Shi and co-workers proposed a pH-driven interlocked DNA nano-spring for the specific stimulation of T-cell proliferation under a slightly acidic tumor microenvironment (Fig. 3c).62 By encoding a cytosine-rich i-motif structure in interlocked circular DNA nanostructures, a spring-like shrinking was achieved in response to the relatively low pH value in tumors to regulate the nanoscale distribution of T-cell receptors (CD3) on the cell surface. This platform elicited significant T-cell proliferation and subsequently enhanced the antitumor effect. This report provided a general strategy for the smart manipulation of various receptors on cell membranes.
Nuclease-responsive strategy
Ribonuclease H (RNase H) and Dicer are two representative nucleases used in nuclease-responsive systems for gene therapy.65 RNase H hydrolyzes the phosphodiester bonds of the RNA strand in DNA/RNA hybrids without exerting a cleavage effect towards the DNA strand.66 Using this feature, gene therapeutic drugs have been rationally incorporated into nuclease-responsive nucleic acid nanostructures for efficient delivery and controllable release.By utilizing roll circling amplification, Gu and co-workers synthesized the yarn-like DNA nanoclews to co-deliver single-guide RNA (sgRNA) and Cas9 protein for genome editing (Fig. 4a).67 The sgRNA/Cas9 complex was loaded on DNA nanoclews containing a partially complementary sequence to sgRNA. The co-assembled complex was subsequently coated with polyethylenimine (PEI) to enhance the following endosomal escape probability. This RCA-based strategy provided a facile method to deliver other nucleic acid drugs or DNA-binding proteins. Assisted by a polymer chain serving as the framework, Zhang and co-workers reported a crosslinked nucleic acid nanogel for siRNA delivery (Fig. 4b).68 The crosslinked nanogel was co-assembled by multivalent DNA-grafted polycaprolactone (DNA-g-PCL) brushes as the framework and functional siRNAs (siRNA-L) with single-stranded overhangs as cross-linkers. The siRNA-L constitutes a double-strand siRNA and two overhangs for base pairing with the DNAs grafted on PCL. Thus, when the nanogel enters the cells, RNase H can specifically recognize and degrade the RNA parts of the DNA/RNA hybrids in the nanogel for the controllable release of siRNA. The formed nanogel exhibited good thermostability and resistance to enzymatic degradation to improve the delivery efficiency of siRNA. In 2021, Ding and co-workers introduced another co-assembled nanoplatform consisting of branched antisense DNA and siRNA for combined gene silencing of target mRNA at different recognition sites (Fig. 4c).69 siRNA with the 3′ overhangs can be efficiently captured by one pair of branched antisenses through DNA/RNA hybridization. The active targeting group and endosomal escape peptide have been attached to the co-assembled nanosystem by host–guest interactions. After cellular uptake and endosomal escape, the branched antisenses and siRNA can be released by endogenous RNase H digestion. This multifunctional nucleic acid nanoplatform presented a remarkable antitumor effect in vivo based on combined gene silencing (antisense and siRNA) of the tumor-associated gene polo-like kinase 1 (PLK1). This report provided an interesting strategy for in situ combinations of delivery carriers and therapeutic cargos.
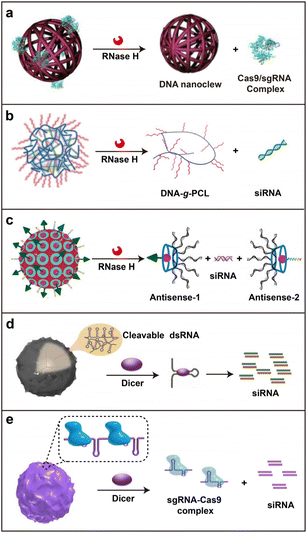 | ||
| Fig. 4 The nuclease-responsive strategy. (a) A DNA nanoclew for CRISPR-Cas9 gene-editing system delivery. Reproduced with permission.67 Copyright 2015, Wiley-VCH GmbH. (b) A crosslinked nucleic acid nanogel for controllable siRNA delivery. Reproduced with permission.68 Copyright 2018, Wiley-VCH GmbH. (c) A co-assembly nanoplatform for the delivery of branched antisense and siRNA. Reproduced with permission.69 Copyright 2021, Wiley-VCH GmbH. (d) A self-assembled RNAi-microsponge based on RCT for shRNA delivery. Reproduced with permission.70 Copyright 2012, Nature Publishing Group. (e) An RCT-based polymeric ribonucleoprotein (RNP) nanoparticle for the co-delivery of CRISPR/Cas9 system and siRNA. Reproduced with permission.71 Copyright 2017, Elsevier. | ||
Differing from the functional mechanism of RNase H, Dicer can process dsRNA into small interfering RNA (siRNA) or microRNA (miRNA) of length 21–23 nt, which can integrate into the RNA-induced silencing complex (RISC) to initiate RNA interference (RNAi) for the intended mRNA degradation.72 Hammond and co-workers reported a self-assembled RNAi microsponge by RCT for shRNA delivery with a high loading capacity and enhanced stability during the delivery process (Fig. 4d).70 After getting access to cells, the tandem shRNA in the microsponge gets cleaved and converted into siRNA by Dicer, resulting in the release of half a million copies of siRNA in each structure for RNAi. Furthermore, Ahn and co-workers integrated both shRNA and the sgRNA/Cas9 complex into an RCT-based nanoparticle (Fig. 4e).71 In response to the intracellular Dicer-mediated cleavage of the shRNA parts, the polymeric ribonucleoprotein nanoparticles produced numerous siRNAs and sgRNA/Cas9 complexes, considerably improving the disruption efficiency of the target gene in cells and animal models. These RCT-based strategies demonstrated the super loading capacity of nucleic acid drugs by their long extension ability.
Biomacromolecule-responsive strategy
Aptamers are single-stranded nucleic acids with high affinity and specificity to bind biomedically relevant biomacromolecules (such as protein) and lower the immunogenicity in vivo compared to antibodies.73 A variety of aptamers, targeting tumor-related biomarkers, have been developed to improve the delivery and internalization efficiency of nanomaterials in cancer therapy.74 With their merits of precise complementary base interaction, easy designability, and chemical modifiability, aptamers have been widely employed for biosensing in the past decades.75,76 Inspired by their specific response towards biomacromolecular triggers, aptamer-based stimuli-responsive nucleic acid nanodevices have been developed.77In 2009, Kjems and co-workers reported a hollow DNA box with a controllable lid functionalized by a “lock-key” system, opening via the strand-displacement reaction.78 Next, Church and co-workers designed a logic-gated DNA nanorobot with a barrel shape for targeted molecular cargo delivery, which was closed by two DNA locks consisting of an aptamer sequence and its corresponding partially complementary strand (Fig. 5a).79 As a proof of concept, they proved that the nanodevice could open after the interaction with antigen keys expressed on the targeted cell membranes, leading to the exposure of the loaded antibody Fab fragments inside to further bind to receptors on the cell surface. Several logical AND-gates were demonstrated by decorating two different kinds of locks. This strategy paved an avenue for constructing specific stimuli-responsive DNA nanodevices for the transportation of active molecular payloads.
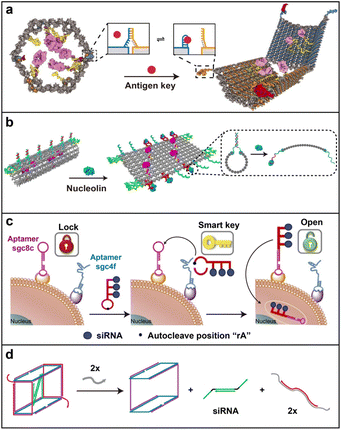 | ||
| Fig. 5 Biomacromolecule-responsive strategy. (a) An aptamer-gated DNA nanorobot for controllable cargo delivery. Reproduced with permission.79 Copyright 2012, American Association for the Advancement of Science. (b) A thrombin-functionalized DNA nanorobot opened by nucleolin binding. Reproduced with permission.80 Copyright 2018, Nature Publishing Group. (c) A “dual lock-and-key” DNA nanotube for cell-subtype-specific and controllable siRNA delivery. Reproduced with permission.81 Copyright 2016, Nature Publishing Group. (d) A DNA nanosuitcase for controlled encapsulation and release of siRNA through strand displacement. Reproduced with permission.82 Copyright 2016, American Chemical Society. | ||
Zhao and co-workers also reported a nucleolin-triggered tubular nanorobot for efficiently transporting thrombin into the tumor site (Fig. 5b).80 In order to realize the stimuli-responsive properties of the nanorobot, six Y-shaped fasteners containing the nucleolin-targeting aptamer (AS1411) were included on the long side of the DNA rectangular origami, serving as the molecular trigger. Additional targeting aptamer sequences were also arranged on both short sides to enhance the targeting effect. After intravenous injection into a tumor-bearing mouse, the tubular DNA nanorobot was opened through recognition by the tumor vessel marker nucleolin, resulting in the exposure of preloaded thrombin inside to activate coagulation at the tumor site for cancer treatment. Briefly, the main concept of this lock–key-responsive strategy was that the bioactive molecules (active drugs) modified by DNA sequence were assembled into the inner cavity or the surface of DNA origami (delivery vehicles), which was locked with DNA aptamer-containing fasteners (locks) and shielded until activation by target biomacromolecules (keys).
To increase the loading capacity of the nanostructures, rolling circle amplification was introduced to the “lock–key” strategy. For instance, Ju and co-workers designed a “dual lock-and-key” for cell-subtype-specific recognition and controllable siRNA delivery (Fig. 5c).81 As the “smart key”, the nano vehicle was modified with an auto-cleavable hairpin structure, which can be activated by two aptamers as “dual locks” sequentially to further mediate the precise delivery of siRNA into specific target cells. This well-defined DNA dual lock-and-key system loaded with siRNA exhibited highly efficient gene silencing with substantially improved delivery specificity and lowered off-target toxicity. Apart from the studies mentioned above, the drug release mechanism based on the strand displacement principle can be broadly included in this category. Representatively, Sleiman and co-workers encapsulated a siRNA in the inner cavity of a self-assembled DNA “nanosuitcase” to protect siRNA against nuclease degradation and unloaded the siRNA in the presence of trigger strands (Fig. 5d).82 An mRNA or microRNA (miRNA) can be utilized as the trigger strand for dual targeting or synergistic therapy in this system. This work reported an interesting attempt at drug release by strand displacement.
Light-responsive strategy
As a noninvasive trigger, light can remotely activate delivery systems to release therapeutic drugs at the targeted position with appreciable spatiotemporal precision and accuracy.83 Normal nucleic acid is irresponsive to light, and thereby, tailored light-responsive molecules need to be rationally designed and introduced.84,85For example, Zhang and co-workers reported a light-triggered DNA–drug nanostructure to efficiently deliver the chemical drug camptothecin (CPT) (Fig. 6a).86 Three hydrophobic CPT molecules were attached to a phenol-based photocleavable linker that subsequently connected with azide-modified DNA strands via the click reaction. The photocleavable properties of the 2-nitrobenzyl group in the linker structure endowed it with a light-responsive ability for DNA–CPT conjugation. Besides, the amphiphilicity of this conjugate drove its self-assembly into micellar nanostructures. Upon a low dose of UV light activation, covalently conjugated CPT was controllably released for chemotherapy. Furthermore, Tan and co-workers prepared an aptamer-grafted photosensitive hyperbranched polymer (HBP) to achieve good biocompatibility, targeting specificity, and controllable drug release (Fig. 6b).87 HBP was functionalized by o-nitrobenzyl moieties and azide groups to realize the UV-responsive properties and connection with DBCO-aptamer, respectively. The conjugated HBP-aptamer self-assembled into stable nanoparticles with a hydrophilic shell and hydrophobic core, whose inner cavities are capable of encapsulating small-molecule drugs (DOX). Under UV irradiation, the nanoparticles dissociated and rapidly released DOX into cancer cells with promising therapeutic effects. These research studies demonstrated that photocleavable bonds could be efficiently exploited for stimuli-responsive drug delivery. However, the employment of UV light limited further applications of these light-triggered systems for drug delivery in vivo due to the low tissue penetration and toxicity of UV light.
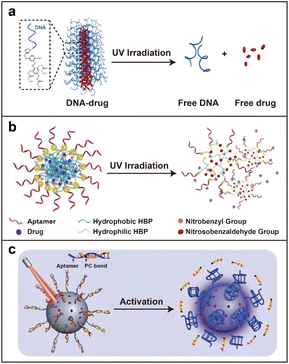 | ||
| Fig. 6 Light-responsive strategy. (a) A light-triggered DNA–drug nanostructure for efficient delivery of the chemo-drug. Reproduced with permission.86 Copyright 2015, American Chemical Society. (b) An aptamer-grafted photosensitive hyperbranched polymer for DOX delivery. Reproduced with permission.87 Copyright 2018, Wiley-VCH GmbH. (c) NIR light-controlled DNA nanodevice for biorecognition and tumor treatment. Reproduced with permission.88 Copyright 2020, American Association for the Advancement of Science. | ||
To avoid these drawbacks of UV light, upconversion nanoparticles (UCNPs) can be employed in light-responsive systems as they can absorb near-infrared (NIR) light and emit UV light. Li and co-workers successfully developed a NIR-light-activated immune device to remotely control antitumor immunity in vitro and in vivo without apparent systemic toxicity.89 A year later, Li and co-workers introduced a DNA nanodevice regulated by orthogonal NIR light for precise tumor recognition and treatment with enhanced spatiotemporal resolution (Fig. 6c).88 The nanodevice was built by incorporating UV light-triggerable aptamer modules and photosensitizers on the surface of the UCNP. After irradiation by two NIR lights with distinct wavelengths, the UCNPs can emit orthogonal UV and green up-conversion luminescence for programmable photoactivation of the aptamer modules and photosensitizers, respectively. This work provided an efficient strategy for spatiotemporally controlled biorecognition and photodynamic antitumor therapy.
Conclusions and perspectives
Over the past decades, versatile nucleic acid nanostructures have been developed and successfully employed for biomedical applications. In this minireview, we have summarized the recent advances in stimuli-responsive nucleic acid nanostructures for efficient drug delivery. Nucleic acid nanostructures can be easily modified with various stimuli-responsive elements, such as disulfide linkages, DNA triplex, i-motifs, targeting aptamers, photocleavable bonds, and so on. After tailored modifications, five representative stimuli-responsive strategies for drug delivery, including redox gradients, pH, nucleases, biomacromolecules, and light, were highlighted. Owing to their good biocompatibility, structural programmability, spatial addressability, and convenient chemical modifiability, stimuli-responsive nucleic acid nanostructures have exhibited remarkable performances in the field of stimuli-triggering drug delivery systems.However, several challenges still remain for these stimuli-responsive nucleic acid nanoplatforms for further practical applications in the clinic. First, systemic pharmacokinetics (absorption, distribution, metabolism, and excretion) need more attention. In particular, their mechanisms of metabolism and excretion in vivo are unclear and need systematic investigation. Second, biosafety is another concern. When a high dosage of the nucleic acid nanostructure is administered, potential immunogenicity stimulation needs to be considered. Third, the scale-up production of nucleic acid nanostructures is still a vital step in reaching the clinical stage. Facile and mild modification and preparation processes with low-cost and time-saving virtues are desired to produce nucleic acid nanostructures with high yield and quality.
In spite of the above-mentioned challenges, nucleic acid nanostructures with inherent advantages are still regarded as the most promising carriers for drug delivery. To date, a series of chemical modification strategies have been developed to enhance the stability of nucleic acids. Fortunately, the immunogenicity of nucleic acid nanostructures can be greatly reduced by rational sequence designs. Meanwhile, the liquid-phase synthesis of nucleic acids may be a promising solution for the scale-up production of nucleic acid nanostructures. We envision that these smart and stimuli-responsive nucleic acid nanostructures will be widely utilized in pre-clinical and clinical studies in the near future.
Author contributions
C. Y. and X. W. contributed equally. J. L. and B. D. conceived the idea and initiated the project. C. Y. and X. W. mainly wrote the manuscript and reproduced the figures. J. L. and B. D. revised the manuscript. All authors confirmed the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Key R&D Program of China (2021YFA1200302 and 2018YFA0208900), the National Natural Science Foundation of China (22025201, 22077023, and 21721002), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB36000000), the CAS Project for Young Scientists in Basic Research (YSBR-036), the CAS Interdisciplinary Innovation Team, the Youth Innovation Promotion Association CAS, and the K. C. Wong Education Foundation (GJTD-2018-03).References
- S. Mura, J. Nicolas and P. Couvreur, Nat. Mater., 2013, 12, 991 CrossRef CAS PubMed
.
- V. P. Torchilin, Nat. Rev. Drug Discovery, 2014, 13, 813 CrossRef CAS PubMed
.
- E. Ruiz-Hernández, A. Baeza and M. Vallet-Regí, ACS Nano, 2011, 5, 1259 CrossRef PubMed
.
- M. Vázquez-González and I. Willner, Angew. Chem., Int. Ed., 2020, 59, 15342 CrossRef PubMed
.
- S. Merino, C. Martin, K. Kostarelos, M. Prato and E. Vazquez, ACS Nano, 2015, 9, 4686 CrossRef CAS PubMed
.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459 CrossRef CAS PubMed
.
- Y. Li, W. Xiao, K. Xiao, L. Berti, J. Luo, H. P. Tseng, G. Fung and K. S. Lam, Angew. Chem., Int. Ed., 2012, 51, 2864 CrossRef CAS PubMed
.
- J. Zhuang, M. R. Gordon, J. Ventura, L. Li and S. Thayumanavan, Chem. Soc. Rev., 2013, 42, 7421 RSC
.
- R. J. Dong, Y. F. Zhou, X. H. Huang, X. Y. Zhu, Y. F. Lu and J. Shen, Adv. Mater., 2015, 27, 498 CrossRef CAS PubMed
.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101 CrossRef CAS PubMed
.
- L. He, J. Mu, O. Gang and X. Chen, Adv. Sci., 2021, 8, 2003775 CrossRef CAS PubMed
.
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237 CrossRef CAS PubMed
.
- N. R. Kallenbach, R.-I. Ma and N. C. Seeman, Nature, 1983, 305, 829 CrossRef CAS
.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Science, 2005, 310, 1661 CrossRef CAS PubMed
.
- P. W. K. Rothemund, Nature, 2006, 440, 297 CrossRef CAS PubMed
.
- L. H. Eckardt, K. Naumann, W. Matthias Pankau, M. Rein, M. Schweitzer, N. Windhab and G. von Kiedrowski, Nature, 2002, 420, 286 CrossRef CAS PubMed
.
- K. V. Gothelf, A. Thomsen, M. Nielsen, E. Cló and R. S. Brown, J. Am. Chem. Soc., 2004, 126, 1044 CrossRef CAS PubMed
.
- J. Zimmermann, M. P. J. Cebulla, S. Mönninghoff and G. von Kiedrowski, Angew. Chem., Int. Ed., 2008, 47, 3626 CrossRef CAS PubMed
.
- Y. Wang, X. Lu, X. Wu, Y. Li, W. Tang, C. Yang, J. Liu and B. Ding, Innovation, 2022, 3, 100217 CAS
.
- Y. Dong, C. Yao, Y. Zhu, L. Yang, D. Luo and D. Yang, Chem. Rev., 2020, 120, 9420 CrossRef CAS PubMed
.
- J. B. Lee, S. Peng, D. Yang, Y. H. Roh, H. Funabashi, N. Park, E. J. Rice, L. Chen, R. Long, M. Wu and D. Luo, Nat. Nanotechnol., 2012, 7, 816 CrossRef CAS PubMed
.
- J. Wang, H. Wang, H. Wang, S. He, R. Li, Z. Deng, X. Liu and F. Wang, ACS Nano, 2019, 13, 5852 CrossRef CAS PubMed
.
- J. Wang, S. Yu, Q. Wu, X. Gong, S. He, J. Shang, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2021, 60, 10766 CrossRef CAS PubMed
.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2008, 47, 6330 CrossRef CAS PubMed
.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D.-K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324 RSC
.
- F. Li, N. Song, Y. Dong, S. Li, L. Li, Y. Liu, Z. Li and D. Yang, Angew. Chem., Int. Ed., 2022, 61, e202116569 CAS
.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607 CrossRef CAS PubMed
.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, J. Am. Chem. Soc., 2012, 134, 1376 CrossRef CAS PubMed
.
- D. Liu, M. Wang, Z. Deng, R. Walulu and C. Mao, J. Am. Chem. Soc., 2004, 126, 2324 CrossRef CAS PubMed
.
- S. Lu, J. Shen, C. Fan, Q. Li and X. Yang, Adv. Sci., 2021, 8, 2100328 CrossRef CAS PubMed
.
- K. R. Kim, S. J. Kang, A. Y. Lee, D. Hwang, M. Park, H. Park, S. Kim, K. Hur, H. S. Chung, C. Mao and D. R. Ahn, Biomaterials, 2019, 195, 1 CrossRef CAS PubMed
.
- C. Y. Tay, L. Yuan and D. T. Leong, ACS Nano, 2015, 9, 5609 CrossRef CAS PubMed
.
- A. T. Veetil, K. Chakraborty, K. Xiao, M. R. Minter, S. S. Sisodia and Y. Krishnan, Nat. Nanotechnol., 2017, 12, 1183 CrossRef CAS PubMed
.
- S. Shi, J. Chen, X. Wang, M. Xiao, A. R. Chandrasekaran, L. Li, C. Yi and H. Pei, Adv. Funct. Mater., 2022, 32, 2201069 CrossRef CAS
.
- M. Madsen and K. V. Gothelf, Chem. Rev., 2019, 119, 6384 CrossRef CAS PubMed
.
- E. Cheng, Y. Xing, P. Chen, Y. Yang, Y. Sun, D. Zhou, L. Xu, Q. Fan and D. Liu, Angew. Chem., Int. Ed., 2009, 48, 7660 CrossRef CAS PubMed
.
- L. Song, V. H. B. Ho, C. Chen, Z. Yang, D. Liu, R. Chen and D. Zhou, Adv. Healthcare Mater., 2013, 2, 275 CrossRef CAS PubMed
.
- Y. Dong, Z. Yang and D. Liu, Acc. Chem. Res., 2014, 47, 1853 CrossRef CAS PubMed
.
- C. Wang, Y. Zhang, Y. Shao, X. Tian, J. Piao, Y. Dong and D. Liu, Giant, 2020, 1, 100006 CrossRef
.
- B. Yang, L. Sun, Y. Pan, Y. Dong, Y. Sun and D. Liu, Acta Polym. Sin., 2021, 52, 996 Search PubMed
.
- Z. Zhao, C. Chen, Y. Dong, Z. Yang, Q.-H. Fan and D. Liu, Angew. Chem., Int. Ed., 2014, 53, 13468 CrossRef CAS PubMed
.
- Y. Li, Y. Ding, B. Yang, T. Cao, J. Xu, Y. Dong, Q. Chen, L. Xu and D. Liu, CCS Chem., 2022 DOI:10.31635/ccschem.022.202101523
.
- B. Yang, Z. Zhao, Y. Pan, J. Xie, B. Zhou, Y. Li, Y. Dong and D. Liu, ACS Appl. Mater. Interfaces, 2021, 13, 48414 CrossRef CAS PubMed
.
- M. Ou, R. Xu, S. H. Kim, D. A. Bull and S. W. Kim, Biomaterials, 2009, 30, 5804 CrossRef CAS PubMed
.
- S. Wang, Y. Zhang, Y. Dong and D. Liu, Polymer, 2019, 175, 146 CrossRef CAS
.
- Y.-N. Zhang, X. Hou, J. Piao, W. Yuan, B.-N. Zhou, X. Zhao, Z. Hao, Y. Zhuang, L. Xu, Y. Dong and D. Liu, Eur. Polym. J., 2022, 173, 111187 CrossRef CAS
.
- G. Saito, J. A. Swanson and K.-D. Lee, Adv. Drug Delivery Rev., 2003, 55, 199 CrossRef CAS PubMed
.
- Y. Lu, W. Sun and Z. Gu, J. Controlled Release, 2014, 194, 1 CrossRef CAS PubMed
.
- J. Li, C. Zheng, S. Cansiz, C. Wu, J. Xu, C. Cui, Y. Liu, W. Hou, Y. Wang, L. Zhang, I.-T. Teng, H.-H. Yang and W. Tan, J. Am. Chem. Soc., 2015, 137, 1412 CrossRef CAS PubMed
.
- J. Zhou, L. Sun, L. Wang, Y. Liu, J. Li, J. Li, J. Li and H. Yang, Angew. Chem., Int. Ed., 2019, 58, 5236 CrossRef CAS PubMed
.
- J. Yang, Q. Jiang, L. He, P. Zhan, Q. Liu, S. Liu, M. Fu, J. Liu, C. Li and B. Ding, ACS Appl. Mater. Interfaces, 2018, 10, 23693 CrossRef CAS PubMed
.
- J. Liu, L. Song, S. Liu, S. Zhao, Q. Jiang and B. Ding, Angew. Chem., Int. Ed., 2018, 57, 15486 CrossRef CAS PubMed
.
- Z. Wang, L. Song, Q. Liu, R. Tian, Y. Shang, F. Liu, S. Liu, S. Zhao, Z. Han, J. Sun, Q. Jiang and B. Ding, Angew. Chem., Int. Ed., 2021, 60, 2594 CrossRef CAS PubMed
.
- H. J. Lee and Y. Bae, Biomacromolecules, 2011, 12, 2686 CrossRef CAS PubMed
.
- E. K. Rofstad, B. Mathiesen, K. Kindem and K. Galappathi, Cancer Res., 2006, 66, 6699 CrossRef CAS PubMed
.
- Z. Zhou, Y. Shen, J. Tang, M. Fan, E. A. Van Kirk, W. J. Murdoch and M. Radosz, Adv. Funct. Mater., 2009, 19, 3580 CrossRef CAS
.
- X. Fu, T. Chen, Y. Song, C. Feng, H. Chen, Q. Zhang, G. Chen and X. Zhu, Small, 2021, 17, 2101224 CrossRef CAS PubMed
.
- Y. Chen, S.-H. Lee and C. Mao, Angew. Chem., Int. Ed., 2004, 43, 5335 CrossRef CAS PubMed
.
- X. Chen, T. Chen, L. Ren, G. Chen, X. Gao, G. Li and X. Zhu, ACS Nano, 2019, 13, 7333 CrossRef CAS PubMed
.
- S. Liu, Q. Jiang, X. Zhao, R. Zhao, Y. Wang, Y. Wang, J. Liu, Y. Shang, S. Zhao, T. Wu, Y. Zhang, G. Nie and B. Ding, Nat. Mater., 2021, 20, 421 CrossRef CAS PubMed
.
- D. Liu and S. Balasubramanian, Angew. Chem., Int. Ed., 2003, 42, 5734 CrossRef CAS PubMed
.
- K. Zhang, Y. Ma, D. Wang, J. Liu, J. An, Y. Li, C. Ma, Y. Pei, Z. Zhang, J. Liu and J. Shi, Nano Lett., 2022, 22, 1937 CrossRef PubMed
.
- Y. Du, P. Peng and T. Li, ACS Nano, 2019, 13, 5778 CrossRef CAS PubMed
.
- R. S. Riley, C. H. June, R. Langer and M. J. Mitchell, Nat. Rev. Drug Discovery, 2019, 18, 175 CrossRef CAS PubMed
.
- R. Kole, A. R. Krainer and S. Altman, Nat. Rev. Drug Discovery, 2012, 11, 125 CrossRef CAS PubMed
.
- W. Keller and R. Crouch, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 3360 CrossRef CAS PubMed
.
- W. Sun, W. Ji, J. M. Hall, Q. Hu, C. Wang, C. L. Beisel and Z. Gu, Angew. Chem., Int. Ed., 2015, 54, 12029 CrossRef CAS PubMed
.
- F. Ding, Q. Mou, Y. Ma, G. Pan, Y. Guo, G. Tong, C. H. J. Choi, X. Zhu and C. Zhang, Angew. Chem., Int. Ed., 2018, 57, 3064 CrossRef CAS PubMed
.
- J. Liu, X. Lu, T. Wu, X. Wu, L. Han and B. Ding, Angew. Chem., Int. Ed., 2021, 60, 1853 CrossRef CAS PubMed
.
- J. B. Lee, J. Hong, D. K. Bonner, Z. Poon and P. T. Hammond, Nat. Mater., 2012, 11, 316 CrossRef CAS PubMed
.
- J. S. Ha, J. S. Lee, J. Jeong, H. Kim, J. Byun, S. A. Kim, H. J. Lee, H. S. Chung, J. B. Lee and D.-R. Ahn, J. Controlled Release, 2017, 250, 27 CrossRef CAS PubMed
.
- L. J. Scherer and J. J. Rossi, Nat. Biotechnol., 2003, 21, 1457 CrossRef CAS PubMed
.
- W. Tan, M. J. Donovan and J. Jiang, Chem. Rev., 2013, 113, 2842 CrossRef CAS PubMed
.
- P. Kumar Kulabhusan and B. Hussain, Pharmaceutics, 2020, 12, 646 CrossRef PubMed
.
- A. D. Keefe, S. Pai and A. Ellington, Nat. Rev. Drug Discovery, 2010, 9, 537 CrossRef CAS PubMed
.
- R. Mo, T. Jiang, R. DiSanto, W. Tai and Z. Gu, Nat. Commun., 2014, 5, 3364 CrossRef PubMed
.
- H.-M. Meng, H. Liu, H. Kuai, R. Peng, L. Mo and X.-B. Zhang, Chem. Soc. Rev., 2016, 45, 2583 RSC
.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. P. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Nature, 2009, 459, 73 CrossRef CAS PubMed
.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831 CrossRef CAS PubMed
.
- S. Li, Q. Jiang, S. Liu, Y. Zhang, Y. Tian, C. Song, J. Wang, Y. Zou, G. J. Anderson, J.-Y. Han, Y. Chang, Y. Liu, C. Zhang, L. Chen, G. Zhou, G. Nie, H. Yan, B. Ding and Y. Zhao, Nat. Biotechnol., 2018, 36, 258 CrossRef CAS PubMed
.
- K. Ren, Y. Liu, J. Wu, Y. Zhang, J. Zhu, M. Yang and H. Ju, Nat. Commun., 2016, 7, 13580 CrossRef CAS PubMed
.
- K. E. Bujold, J. C. C. Hsu and H. F. Sleiman, J. Am. Chem. Soc., 2016, 138, 14030 CrossRef CAS PubMed
.
- G. Shim, S. Ko, D. Kim, Q. V. Le, G. T. Park, J. Lee, T. Kwon, H. G. Choi, Y. B. Kim and Y. K. Oh, J. Controlled Release, 2017, 267, 67 CrossRef CAS PubMed
.
- D. Y. Tam, X. Zhuang, S. W. Wong and P. K. Lo, Small, 2019, 15, 1805481 CrossRef PubMed
.
- C. Wang, M. P. O'Hagan, Z. Li, J. Zhang, X. Ma, H. Tian and I. Willner, Chem. Soc. Rev., 2022, 51, 720 RSC
.
- X. Tan, B. B. Li, X. Lu, F. Jia, C. Santori, P. Menon, H. Li, B. Zhang, J. J. Zhao and K. Zhang, J. Am. Chem. Soc., 2015, 137, 6112 CrossRef CAS PubMed
.
- L. Yang, H. Sun, Y. Liu, W. Hou, Y. Yang, R. Cai, C. Cui, P. Zhang, X. Pan, X. Li, L. Li, B. S. Sumerlin and W. Tan, Angew. Chem., Int. Ed., 2018, 57, 17048 CrossRef CAS PubMed
.
- Z. Di, B. Liu, J. Zhao, Z. Gu, Y. Zhao and L. Li, Sci. Adv., 2020, 6, eaba9381 CrossRef CAS PubMed
.
- H. Chu, J. Zhao, Y. Mi, Z. Di and L. Li, Nat. Commun., 2019, 10, 2839 CrossRef PubMed
.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |
