Open Access Article
Open Access ArticleHoney authenticity: analytical techniques, state of the art and challenges
Aristeidis S. Tsagkaris
 ab,
Georgios A. Koulis
ab,
Georgios A. Koulis
 a,
Georgios P. Danezis
c,
Ioannis Martakos
a,
Marilena Dasenakia,
Constantinos A. Georgiou
c and
Nikolaos S. Thomaidis
a,
Georgios P. Danezis
c,
Ioannis Martakos
a,
Marilena Dasenakia,
Constantinos A. Georgiou
c and
Nikolaos S. Thomaidis
 *a
*a
aLaboratory of Analytical Chemistry, Department of Chemistry, National and Kapodistrian University of Athens, Panepistimiopolis Zographou, 15771 Athens, Greece. E-mail: ntho@chem.uoa.gr; Web: http://trams.chem.uoa.gr/ Fax: +30 210 7274750; Tel: +30 210 7274317
bDepartment of Food Analysis and Nutrition, Faculty of Food and Biochemical Technology, University of Chemistry and Technology Prague, Technická 5, 166 28 Prague 6 – Dejvice, Prague, Czech Republic
cChemistry Laboratory, Department of Food Science and Human Nutrition, Agricultural University of Athens, 75 Iera Odos, 118 55 Athens, Greece
First published on 17th March 2021
Abstract
Honey is a high-value, globally consumed, food product featuring a high market price strictly related to its origin. Moreover, honey origin has to be clearly stated on the label, and quality schemes are prescribed based on its geographical and botanical origin. Therefore, to enhance food quality, it is of utmost importance to develop analytical methods able to accurately and precisely discriminate honey origin. In this study, an all-time scientometric evaluation of the field is provided for the first time using a structured keyword on the Scopus database. The bibliometric analysis pinpoints that the botanical origin discrimination was the most studied authenticity issue, and chromatographic methods were the most frequently used for its assessment. Based on these results, we comprehensively reviewed analytical techniques that have been used in honey authenticity studies. Analytical breakthroughs and bottlenecks on methodologies to assess honey quality parameters using separation, bioanalytical, spectroscopic, elemental and isotopic techniques are presented. Emphasis is given to authenticity markers, and the necessity to apply chemometric tools to reveal them. Altogether, honey authenticity is an ever-growing field, and more advances are expected that will further secure honey quality.
1. Introduction
Honey is a supersaturated sugar solution famous for its beneficial health effects1,2 owing to its highly diverse composition.3 Recent studies showed the honey preventive properties against cancer4,5 and anti-inflammatory value.6 Therefore, an ever-increasing honey consumption has been noticed as a result of a healthy lifestyle that is followed by many individuals. However, this fact generates a supply problem, as many countries around the globe cannot afford a self-sufficient honey production (mostly due to climate conditions and high population), something that forces them to import honey.7 Obviously, honey is a major fraud target in consideration of its price, high consumption and the globalized market,8 and the regulatory requirements vary depending on the country.Honey quality is related both to honey production and origin. The first case is linked to the processing of honey during the production procedure, which includes several steps, for example centrifugation, filtering or pasteurization.9 The European Commission (EC) Directive 2001/110/EC indicates that honey filtering should be stated on the label, but this was not always the case. However, the adulteration of honey with various components during production is a more common problem. This adulteration is profit-driven, as honey is an expensive product featuring a high market price. In this way, cheap sweeteners are incorporated into honey to reduce the amount of actual honey per packaging, with striking examples being high-fructose corn syrup (HFCS), glucose syrup, or invert-sugar syrups.10 Water addition is another less preferred fraudulent practice, which affects the shelf-life and nutritional value.11 Reasonably, analytical methods able to detect honey fraud have been developed, and they have been insightfully described12 and recently discussed.8,13 To this end, this study will not provide further discussion on honey adulteration.
In terms of honey origin, this is a characteristic directly related to the market price and the product quality. The importance of origin is reflected in the European Union (EU) legislation, in which: (i) the necessity to declare honey origin on the label, e.g., “blend of EU honeys”, is emphasized (Directive 2001/110/EC and its amendment Directive 2014/63/EU), and (ii) the establishment of quality schemes is described. In detail, two quality schemes are prescribed in the Regulation (EU) No. 1151/2012, namely “Protected Designation of Origin” (PDO) and “Protected Geographical Indication” (PGI), highlighting the obvious relation between the food quality and geographical origin. Therefore, a genuine honey from a specific region/area/country, for example Mel de Barroso PDO Portuguese honey, can achieve greater market stake and price. A comprehensive list of the PDO and PGI products can be found in the eAmbrosia portal, https://ec.europa.eu/info/food-farming-fisheries/food-safety-and-quality/certification/quality-labels/geographical-indications-register/, (last accessed 18/12/2020), which is the EU geographical indication register. However, honey geographical origin is not the only important authenticity parameter. The botanical origin can also influence the market price. For example, in the EU, there is a trend towards honeydew honey consumption, resulting in a higher market price.14 Overall, it is of paramount importance to monitor and control the honey origin, and this is feasible only with the combination of reliable analytical methods to advanced chemometric tools.
Various honey components, such as physicochemical characteristics, phenolic compounds or metals, have been used as discrimination markers among different botanical and/or geographical origins. These data are generated by various analytical techniques, such as liquid chromatography coupled to mass spectrometry (LC-MS) or nuclear magnetic resonance (NMR), providing a wealth of information. Nevertheless, refining raw data and extracting useful information is a rather challenging task. In fact, the use of advanced chemometric tools is commonly necessary to achieve the desired discrimination. It is common to measure X number of variables in N number of samples to create a matrix Y (X × N) that aims to reveal potential pattern differences that can be exploited either qualitatively (e.g., fingerprinting) or quantitatively (marker compounds indicating a specific botanical origin, such as hotrienol, nerol oxide, and benzyl cinnamate in goldenrod honey15). Multivariate data analysis features both unsupervised methods, such as principal component analysis (PCA) or hierarchical cluster analysis (HCA), and supervised methods such as partial least squares discriminant analysis (PLS-DA) or partial least squares regression (PLSR).16 It is worth noting that in unsupervised methods, the applied algorithm does not have any information regarding the class of the samples, while in the supervised methods, a training set with a known sample classification is necessary in order to generate a predictive model. In addition, there are many published papers with contradictory results depending on the statistical procedure used, indicating the importance of chemometrics in food authenticity.17 To better understand the application of multivariate data analysis and machine learning in honey authenticity studies, a recent review study be C. Maione et al.18 is highly recommended. However, even the most powerful chemometric tools cannot compensate for poor analytical data. Consequently, it is necessary to highlight the trends and discuss the bottlenecks of the available analytical methodologies.
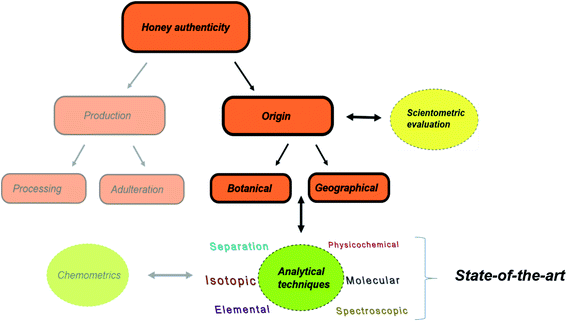
In this review, the state-of-the-art tools related to analytical techniques for honey origin determination are provided (Fig. 1). To depict the ever-increasing research interest on honey authenticity studies, we conducted an all-time scientometric evaluation of the field for the first time. The various analytical methods and applications reported during the period 2014–2019 are critically discussed, providing insight into the authenticity issues, potential markers and applied chemometric tools.
2. Applied methodology to review the available literature
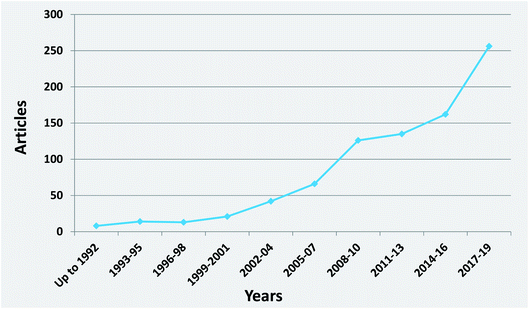
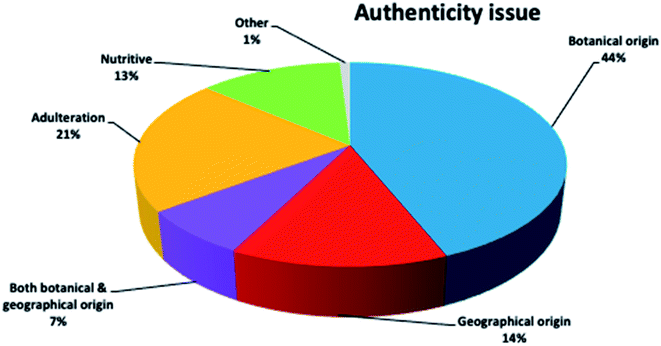
It is interesting to notice the research trends on honey authenticity studies by conducting an all-time scientometric evaluation. The search was performed using the Scopus database and the following search string, “authentication” OR “geographical origin” OR “botanical origin” OR “authenticity” OR “fraud” OR “adulteration” AND honey”. It is important to notice that only research papers written in the English language were included in the study. Furthermore, the remaining papers were evaluated in terms of context to ensure that they were related to honey authenticity. All abstracts (1129 papers, published up to 2019) were analysed for relevance, and all selected articles were comprehensively checked. It was revealed that honey authenticity is a highly evolving field (Fig. 2). Although there is a stabilization of research articles during the years 2011–2013, a significant increase in papers is clearly noticed in the 2017–2019 period, indicating that research interest is still growing in the field.It was also worthwhile to investigate the paper clustering related to the various authenticity issues, namely botanical origin, geographical origin and adulteration (Fig. 3). Botanical origin discrimination was the most studied authenticity issue (about 44% of the reviewed papers), followed by adulteration (about 21%) and geographical origin discrimination (14%). There were also papers investigating both botanical and geographical origin (about 7%). As already stated, honey price is directly connected to its botanical and geographical origin,19 a fact that explains why the majority of papers are focused on honey origin. The search also showed that 13% of the studies dealt with the nutritional value of honey from different botanical or/and geographical origins. There is also a very small percentage (1%) of articles on other authenticity issues, such as production type,20 discrimination between honeydew and blossom honey,21 and discrimination of honeys from different entomological source.22 Importantly, almost all of these articles were published after 2017, pinpointing novel and emergent authenticity issues that will be further investigated in the future.
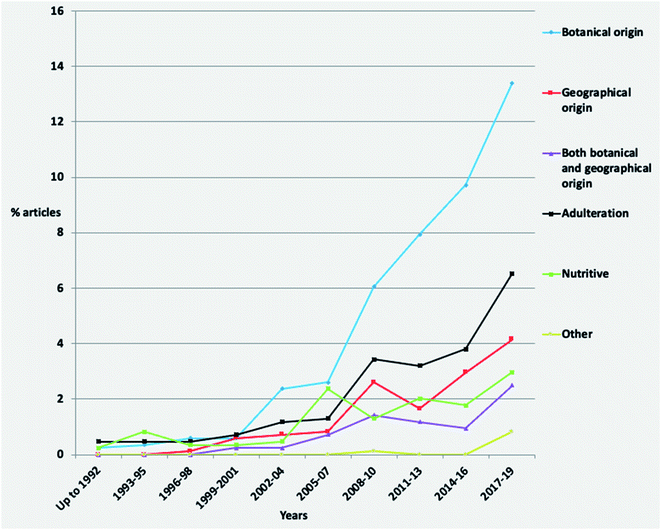
Concerning the temporal evolution of articles per authenticity issue, studies investigating the botanical origin present a high increase since the 2008–2010 period (Fig. 4). Adulteration and geographical origin articles show almost a stable increase, while the percentage of nutritive studies has been the same since 2005–2007. The last could be attributed to the generally well-defined nutritional value of honey.
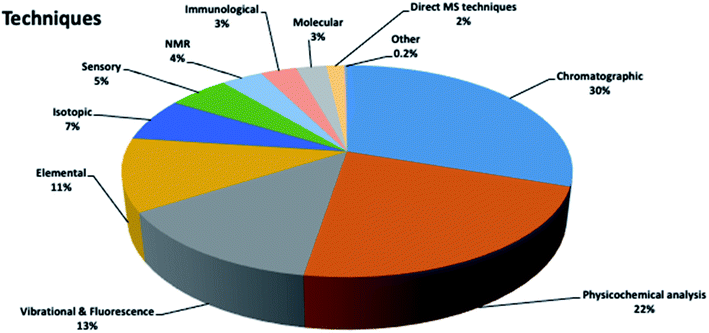
Honey authenticity was studied using a variety of analytical techniques (Fig. 5). Interestingly, more than 50% of the articles used chromatographic techniques (30%) and physicochemical analysis (22%) for the determination of honey physicochemical properties (pH, electrochemical conductivity, color, viscosity). In the case of chromatographic techniques, although they are expensive, complicated and time-consuming, they are considered powerful tools that are able to achieve impressive discriminant results when combined with chemometrics (see Section 3.2). On the other hand, physicochemical analysis is usually the first step when trying to monitor honey origin, as it is low-cost, fast and simple (see Section 3.1). Subsequently, vibrational and fluorescence spectroscopy, and elemental, isotopic, sensory, NMR, immunological and molecular techniques were used.
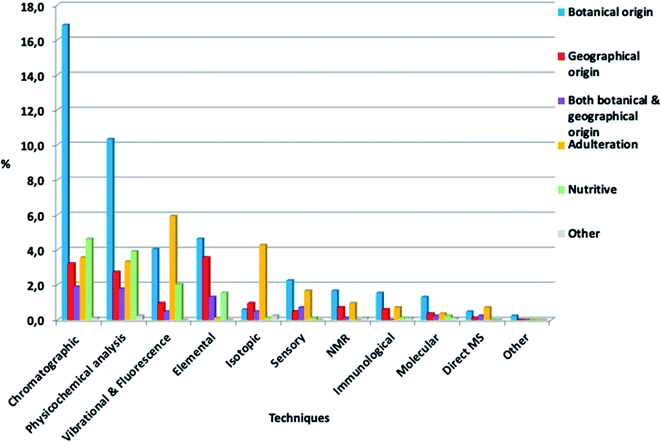
It is also interesting to notice which techniques were mostly studied for each authenticity issue (Fig. 6). Chromatographic techniques and physicochemical analysis were mainly used for botanical origin determination studies. Vibrational and fluorescence techniques were mostly applied to reveal adulteration cases, as well as botanical origin determination studies. In terms of immunological and molecular techniques, they were mostly used for botanical origin determination, as expected, since these techniques have been utilized for genetic origin determination. Isotopic techniques were mostly used in adulteration studies. Specifically, stable carbon isotope ratio analysis (SCIRA) is the official method for the detection of honey adulteration, and it is based on the stable carbon isotope ratio, specifically 13C/12C = δ13C (‰). This ratio can reveal sugar addition in honey, as well as bees overfeeding.
3. Analytical techniques and applications
To highlight the current situation on analytical techniques used in honey authenticity studies, we critically reviewed the available literature based on the collected scientometric data of the last six years (2014–2019). Pollen analysis or melissopalynology, the pollen investigation under a microscope to reveal honey origin, is the traditional way to confirm honey origin. However, it is not included in the following paragraphs, as this approach does not reflect the current state-of-the-art approaches.23 Although it can provide reliable results and is sometimes used to benchmark the results obtained by modern instrumental analysis, melissopalynology needs experienced personnel and the sample throughput is low.24 In terms of the discussed analytical techniques, they determine different honey characteristics and can be exploited differently in the various authenticity issues. Tables containing authenticity markers for each technique, alongside the utilized chemometric tools, are presented and summarize very useful information, which can be highly beneficial for future authenticity studies.3.1 Physicochemical and sensory analysis
Certain honey physicochemical characteristics are prescribed in the EU legislation as the quality criteria that should be met by a product. These characteristics include sugars, moisture and water insoluble content, electrical conductivity, free acidity, hydroxymethylfurfural (HMF) and diastase activity. The analysis methods for these parameters have been established by the International Honey Commission (IHC),25 including both instrumental, such as HPLC for sugars or HMF determination, and classical analysis such as reflectometry or titrations. Physicochemical characteristics have been exploited to investigate honey authenticity as the simplest and least expensive approach, indicating the need for conventional methods to assure honey authenticity (Table 1). To begin with, physicochemical parameters were used to successfully discriminate the geographical origin of 141 honey samples from 5 different Argentinian regions.26 Interestingly, the determination of just five parameters, i.e., moisture, electrical conductivity, pH, acidity and HMF, coupled to PCA and linear discriminant analysis (LDA) achieved a 65.8% correct classification among the 5 regions. However, when two specific regions were selected (Corrientes and Formosa provinces), the correct classification was increased up to 98.7%. Physicochemical parameters were also used as authenticity markers for the botanical discrimination of Polish honey.27 In this case, 72 samples from eight different botanical species were correctly characterized with a 99% accuracy by applying chemometrics, specifically, classification and regression trees (C&RT).| Authenticity issue | Markers | Method | Chemometric tool | Ref. |
|---|---|---|---|---|
| Botanical discrimination (17 unifloral honeys from Europe) | Color coordinates: L*, a*, b*, c*ab, h*ab | CIE L C chromaticity coordinates using a UV-Vis spectrophotometer | HCA did not provide discrimination, correct prediction rate was not stated for PCA | 33 |
| Botanical discrimination (pine, thyme, fir, orange blossom) | 4 phenolic compounds (quercetin, syringic acid, kaempferol, myricetin) and 10 conventional quality parameters | Physicochemical characteristic measurement + HPLC-DAD | Multivariate Analysis Of Variance (MANOVA) + LDA, using: 4 phenolic compounds and 10 conventional quality parameters 96.6% correct prediction | 28 |
| Geographical discrimination of Argentinian honey (5 regions) | Moisture, electrical conductivity, pH, acidity and HMF | 9 physicochemical parameters | PCA + LDA: 65.8% for samples originating from 5 regions, 98.7% correct prediction for samples from 2 different regions | 26 |
| Botanical discrimination | Pollen and physicochemical properties | Palynological and physicochemical analysis | Cluster analysis (CA) and PCA | 29 |
| Honeydew vs. blossom | Color | Colorimeter | PCA and classification and regression trees (C&RT) | 34 |
| Botanical discrimination (8 botanical species from Poland) | Physicochemical properties | Physicochemical char. measurement | C&RT, 99% correct prediction | 27 |
| Geographical characterization of Italian honey | Color and total antioxidant capacity | Optical comparator and spectrophotometer for color, ABTS and 2,2-diphenyl-1-picrylhydrazyl (DPPH) assays for antioxidants | HCA and PCA | 35 |
| Botanical origin (Eucalyptus camaldulensis vs. Myrtus communis) | Combination of physicochemical, sensory and pollen data | Physicochemical char measurement, sensory, pollen | CA and PCA | 30 |
| Italian multifloral vs. unifloral honey | Sensory characteristics and pollen | e-tongue and melissopalinology | 100% match between the two techniques | 36 |
| Botanical discrimination (acacia, sunflower, tilia, honeydew, and polyfloral) | Electrochemical data + physicochemical properties (pH, free acidity, electrical conductivity) | Physicochemical measurements + voltammetric electronic tongue + HPLC | PCA classification and LDA for electrochemical data, PLS to reveal correlations to physicochemical properties | 37 |
There were also cases in which physicochemical characteristics were combined with other analytes such as volatile compounds, phenolic compounds or elements, to build more reliable and accurate prediction models. In this way, 4 phenolic compounds (quercetin, syringic acid, kaempferol, myricetin) and 10 conventional quality parameters were combined in multivariate models (LDA) to differentiate Greek pine, thyme, fir, orange blossom honey, achieving 97% accuracy.28 Elements were also used to assist both geographical and botanical differentiation, e.g., Na, Mg, K, Ca, Mn, Zn. These elements, alongside the total dissolved sugar, electrical conductivity, acidity, total ash and color, were revealed to be Saudi Arabian honey markers using the HCA, PCA and stepwise discriminant analysis (DA). In conclusion, although the application of instrumental methods resulted in an increased analysis cost, the determination of physicochemical characteristics was not always enough to define honey origin.29
Sensory analysis is an alternative approach, usually combined with physicochemical parameters determination, in authenticity studies. The unique organoleptic honey characteristics can be exploited to indicate the floral origin. Obviously, this method requires a well-trained panel and statistical evaluation of the results. A striking example of this can be found in the study by I. Rodríguez et al.,30 in which highly experienced assessors differentiated unifloral Spanish honeys assisted by melissopalynological data. However, the major drawback of sensory analysis, even based on trained and experienced individuals, is the inherent lack of standardization. To counter this limitation, the so-called electronic tongues (e-tongues) or electronic noses (e-noses) have emerged. These methods employ non-specific sensor-arrays mimicking human senses. For instance, a potentiometric e-tongue was successfully applied to differentiate honeys (n = 67) of different floral origin (Castanea sp., Echium sp., Erica sp., Lavandula sp., Prunus sp. and Rubus sp.).31 Initially, honey samples were grouped based on their color (white, amber and dark) and 100% classification accuracy was achieved, following the leave-one-out (LOO) cross validation approach. Similarly, an e-nose provided non-destructive analysis, predicting the physicochemical characteristics based on the aroma profile.32 The prediction was based on the maximum value of the response time and the area under the response time curve processed by an artificial neural network (ANN). To sum up, sensory analysis can provide complementary information to physicochemical properties, and artificial sensing has the potential to enhance the rapidness and cost-efficiency in food authenticity studies.
3.2 Separation techniques
Separation techniques are considered powerful analytical tools, which have been widely used in the food authenticity field. Their main advantage is that they can separate analytes of interest from complex food matrices, enhancing the method detectability, accuracy and precision. As it is depicted through the performed scientometric analysis (see “2. Applied methodology to review the available literature”), chromatographic methods are used the most to determine honey authenticity. Liquid chromatography (LC) is more frequently used as it can measure a wider number of compounds, in contrast to gas chromatography (GC), which can only detect thermostable (semi-)volatile analytes. Electrophoresis, another separation method able to cluster proteins based on their molecular weight, has not been extensively used in honey authenticity. The potential to utilize electrophoresis for entomological source discrimination was demonstrated in a study by Ramon-Sierra et al.,38 in which stingless bee honey from Apis milifera honey was classified by combining electrophoresis to the physicochemical characteristic measurement. In the following paragraphs, expert opinion on the LC and GC methods used in honey authenticity studies is provided.| Authenticity issue | Markers | Method | Chemometric tool | Ref. |
|---|---|---|---|---|
| Geographical origin discrimination of Serbian polyfloral honeys | Chlorogenic acid, ellagic acid, quercetin, dicaffeoylquinic acid, pinobanksin 5-methylether-3-O acetate, bis-methylated quercetin, pinobanksin 3-O-propionate, pinocembrin, galangin, eriodictyol, sakuranetin, pinobanksin, acacetin, caffeic acid phenethyl ester, rhamnetin, caffeic acid, isorhamnetin, and methoxychrysin | UHPLC-LTQ OrbiTrap MS + total phenolic content & radical scavenging activity assays | Kruskal–Wallis test, PCA (5 PCs explained 68.99% of the variance) and PLS-DA | 51 |
| Geographical origin discrimination of Greek thyme honey | Chrysin, syringic acid, quercetin, kaempferol and myricetin | HPLC-UV + physicochemical parameters | MANOVA and LDA (two discriminant functions explained 94.1% of total variance), overall correct classification 83.3% | 48 |
| Botanical classification (chaste, rape) | Kaempferol, morin, ferulic acid | HPLC-DAD-MS/MS | PCA (2 PCs explained 64.83% of the variance), PLS-DA and SIMCA. Discrimination accuracy for calibration set was 94.53% and for predictive set 96.43% | 52 |
| Authenticity of unifloral sage honey | Boron, potassium, kaempferol, turanose | UHPLC DAD-MS/MS, UHPLC-LTQ OrbiTrap MS + total phenolic content, radical scavenging activity assays, HPAEC-PAD & ICP-OES | PCA (2 PCs explained 32.18% of the variance) | 44 |
| Botanical discrimination of New Zealand honeys (manuka, clover kamahi, rata) | Non-targeted markers with potential structures | UPLC-QToF/MS + IRMS, ICP-MS & Vibrational spectroscopy | PCA, Orthogonal partial least square discriminant analysis (OPLS-DA) | 50 |
| Characterisation of phenolic compounds in Algerian honeys and botanical discrimination among 12 species | Caffeic acid for Capparis spinosa honey and p-coumaric acid for Trifolium and Annarhinum honeys | HPLC-ESI-TOF-MS + pollen analysis, total phenolic content & total flavanoid content | ANOVA | 53 |
| Floral (leatherwood and meadow, manuka, kamahi) and geographical origin (Australia, New Zealand, others) discrimination | 6a-Dihydrocornic methyl ester-60-O-b-D-glucopyranoside, leptosperin, phenyllactic acid, 4-methoxyphenyllactic acid, pinobanksin, dihydroxyisoflavone, methyl syringate, homovannilic acid, 2-phenylethyl b-D-glucopyranoside, syringic acid, gallic acid, hydrocinnamic acid, pinobanskin, 4-methoxycinnamic acid, eugenic acid and unknowns | UPLC-QToF MS | PCA, OPLS-DA | 54 |
| Floral (Litchi, acacia) and geographical origin (regions of China) discrimination | Geranial, (±)abscisic acid, 10-HAD, abscisic aldehyde, naringenin chalcone, abscisic acid glucose ester, paeonoside, cynaroside A, hypaphorine, 3,4-dimethoxycinnamic acid, alpha-curcumene, malic acid, beta-cyclocitral, 4-methylcinnamic acid, pinobanksin, cinnamyl alcohol, cinnamic acid, isosakuranetin, L-aspartic acid, beta-cyclocitralin, hydroxycinnamic acid, luteolin and Boc-D-tyrosine | UHPLC-Q-Orbitrap | PCA, Volcano plot, VIP | 42 |
| Botanical discrimination of Romanian honeys (acacia, tilia, sunflower, honeydew, polyfloral) | Not available | HPLC-DAD + physicochemical analysis | PCA, LDA (correct classification 92.0%), ANN (correct classification 94.8%) | 55 |
| Authenticity of orange honey | Synephrine | LC-ESI-MS/MS + pollen analysis | — | 45 |
| Botanical origin of Australian honey | Pyrrolizidine alkaloids, lycopsamine, indicine and intermedin | UHPLC-MS/MS | — | 46 |
| Discrimination of entomological source Sicilian black honeybees (Apis mellifera ssp. sicula) and common honeybees (Apis mellifera ssp. ligustica) | Kaempferol, quercetin, myricetin, pinocembrin, caffeic acid and chlorogenic acid | LC–ESI-Orbitrap™-MS/MS | PCA | 56 |
| Determination of organic acids in commercial honey samples using stable carbon isotope ratios | Gluconic acid | LC/IRMS | Descriptive statistics | 57 |
| Investigation of geographical origin discrimination (7 countries) | No significant differences in the content of gluconic acid | LC/IRMS | Descriptive statistics | 58 |
| Honeydew honey characterization and discrimination to blossom honey | Quercetin, naringenin, caffeoylquinic acid, hydroxyphenylacetic acid, apigenin and genistein | UHPLC-LTQ OrbiTrap MS, UHPLC-DAD-MS/MS + total phenolic content, radical scavenging activity assays & cyclic voltammetry | Descriptive statistics of variance, Kruskal-Wallis one-way analysis, PCA | 49 |
| Botanical origin discrimination (lime and acacia honey) | 7 phenolic compounds and abscisic acid | HPTLC | PCA (2 PCs explained 54% of the variance) | 47 |
The use of various analytical detectors hyphenated to LC is also reported (Table 2). Of importance is to highlight that honey authenticity is feasible even by using simple analytical methods, such as high performance thin layer chromatography (HPTLC)47 or LC coupled to an ultraviolet (UV) detector.48 In the first case, 7 phenolic compounds and abscisic acid were used as markers to discriminate between lime and acacia honeys based on PCA classification. In contrast, in the latter study, the geographical origin of Greek thyme honey was differentiated by combining chrysin, syringic acid, quercetin, kaempferol and myricetin content to honey quality parameters, such as free acidity or color values. In terms of MS-based detection, both low- and high-resolution mass spectrometry (LRMS and HRMS) methods have been employed to investigate for honey authenticity. In fact, there were cases combining targeted and non-targeted analysis using LRMS and HRMS, respectively. In this way, ultra-high-performance liquid chromatography coupled with a hybrid mass spectrometer (UHPLC-LTQ OrbiTrap MS) and ultra-high-performance liquid chromatography with a diode array detector and a triple-quadrupole mass spectrometer (UHPLC-DAD-MS/MS) were combined for studying the Croatian honeydew honey, identifying 52 compounds and quantifying 25 of them.49 Obviously, the screening capabilities provided by HRMS in the full scan mode can be complementary to the wide linear ranges, low detection limits, and excellent accuracy of the triple quadrupole MS. Another promising approach was the hyphenation of an LC system to isotope ratio mass spectrometry (LC/IRMS) in order to measure for organic acids. The LC part consisted of two columns. Initially, a size exclusion column was necessary to separate the carbohydrates and organic acids due to their similar chemical composition. Then, organic acids were separated from each other on a RP system and analyzed by IRMS. The potential to use gluconic acid as a geographical origin indicator (from 7 different countries) was investigated, but more evidence is needed to ascertain this. Last but not least, although LC based methods acquired a wealth of analytical data, there were still cases in which data fusion with other analytical methods, e.g., vibrational spectroscopy or elemental analysis, can provide additional authenticity markers and more robust prediction models.50
| Authenticity issue | Markers | Method | Chemometric tool | Ref. |
|---|---|---|---|---|
| Geographical discrimination of acacia, sunflower and tilia honey (Spain, Romania, and the Czech Republic) | 2-Methyl-2-butenal and 2-methyl-2-propanol for acacia honeys; 1-hexanol and α-pinene for sunflower honeys and 3-methyl-1-butanol and hotrienol for tilia honey | P&T-GC/MS + sugars + physicochemical parameters | ANOVA, PCA, SLDA (classification of 100% of acacia and tilia honeys and 93.8% of sunflower) | 68 |
| Geographical origin discrimination of Greek thyme honey (Irakleio, Hania, Kefalonia, Symi, Lakonia) | Formic acid ethyl ester, formic acid, acetic acid, 1-hydroxy-2-propanone, octane, terpinen-4-ol, decanal, decanoic acid ethyl ester and 4,7,7-trimethyl-bicyclo [3,3,0]-octan-2-one + 11 physicochemical parameters | HS-SPME-GC/MS + physicochemical parameters | MANOVA and LDA (two discriminant functions explained 92.2% of total variance), overall correct classification 92.9% | 59 |
| Geographical origin discrimination of Greek pine honey (Halkidiki, Evia, Thassos, Samos) | Hexanoic acid ethyl ester, 2,3 butanediol, decane, beta-thujone, heptanoic acid ethyl ester, 1-methyl-4-(1-methylethenyl)benzene, nonanal and 2-ethyl-1-hexanol + 9 physicochemical parameters | HS-SPME-GC/MS + physicochemical parameters | MANOVA and LDA (three discriminant functions explained 98% of total variance), overall correct classification 74.4% | 60 |
| Gotanical discrimination of Greek unifloral honeys (pine, thyme, fir, orange) | 30 volatile compounds (among other, 1-hydroxy-2-propanone and decane for thyme honey; 6-methyl-5-hepten-2-one and 2-hydroxy-3,5,5-trimethylcyclohex- 2-en-one for fir honey; beta-thujone for pine honey; linalool, (E)-linalool oxide, limonene for orange honey) + 10 physicochemical parameters | HS-SPME-GC/MS + physicochemical parameters | Multi dimensional one-way analysis of variance (MANOVA) and LDA (two discriminant functions explained 98.3% of total variance), overall correct classification 95.8% | 61 |
| Geographical origin discrimination (Galician, Malaysia, Bangladesh) of mono and multi-flora honeys | Toluene | HS-SPME-GC-QTOF-MS | PCA (2 PCs explained 92% of the variance) | 74 |
| Authenticity of orange honeys | Linalool, linalool oxide isomers | HS-SPME-GC-ion trap | — | 73 |
| Botanical origin discrimination (heather, raspberry, rape, alder buckthorn) | Benzoic acid, isophorone, 2-methylbutyric acid and absent of linalool for heather honey | HS-SPME-GC-TOF | Hierarchical cluster analysis and correspondence analysis | 75 |
| Geographical classification of bracatinga honeydew honey | Free amino acid profile | GC-MS | Cluster analysis (CA), PCA (2 PCs account for 82%) | 78 |
| Label verification for Levanter and thyme honeys | Ethyl acetate, 2,3-butanedione, 2-methylpropanenitrile and 1-butanol for thyme honey; 1-hexanol, hotrienol, hexanal, acetic acid and 2-methyl-2-buten-1-ol for levander honey | P&T-GC/MS + physicochemical parameters + sensory analysis | PCA (7 PCs explained 95.5% of the variance), SLDA (one discriminant function explained 100% of total variance), correct classification 85.7% | 79 |
| Geographical (regions of Brazil) and entomological (8 species of stingless bees) origin discrimination | Ethyl octanoate, ethyl decanoate, hotrienol, epoxylinalol, benzaldehyde, TDN, thuja-2,4 (10)-diene, ethyl hexanoate, p-cymene, 2-heptanol and 2-heptanone | HS-GC-MS | PCA (6 PCs explained 91.9% of the variance), LDA, correct classification 100% | 67 |
| Authenticity of sugarcane honey (certified, non-certified) | 1,2,3,4-Tetrahydro-1,1,6-trimethyl-naphthalene and acetic acid for certified; 1-methyl-2- pyrrolidinone, 1-methyl-4-(1-methylethyl)-benzene, 1,2,3,4-tetrahydro-1,5,8-trimethyl-naphthalene and 4,6-dimethyl-pyrimidine for non-certified | HS-SPME-GC/MS | One-way ANOVA, PCA (2 PCs account for 86.1%), LDA (classification rate of 100%) | 62 |
| Botanical (angico, algaroba, chanana, malícia) and entomological (urucu, jandaíra) origin discrimination | Furans and aromatic aldehydes for urucu/angico; sulphur compounds for jandaíra/algaroba; terpenes, norisoprenoids, esters, alcohols and hydrocarbons for urucu/chanana and jandaíra/chanana; ketones for urucu/malicia and jandaíra/malicia | HS-SPME-GC/MS | PCA (2 PCs explained 71% of the variance) | 65 |
| Botanical discrimination of European honeys (acacia, canola, honeydew) | Hexanal, cis-linalool oxide, benzaldehyde,3-hydroxy-2-butanone (acetoin), trans-2-pentenal, and 3-methylbutanol | HS-GC-IMS | PCA (10 PCs explained >90% of the variance), LDA (overall classification of 98.6%), kNN (overall classification of 86.1%) | 71 |
| Geographical discrimination of Acacia honey from Romania | 2-Butanol, 5-methyl-2-hexanone, 2-heptanone, octanal, 2,2-dymethyl propanoic acid, naphthalene, nonanoic and octanoic acids, borneol and HMF, pentanoic acid | HS-SPME-GC-MS + physicochemical parameters | One-way ANOVA | 63 |
| Geographical discrimination of Algerian honeys (Arid and Mediterranean Areas of Algeria) | 1,3-Di-tertbutylbenzene for thyme honey; mesitylene for Solidago canadensis L. honey; decane fo rhododendron, chestnut and honeydew honey; 1-methoxy-4-methylbenzene for Salix spp. honey; isophorone for heather honey | HS-SPME-GC/MS | Ascending hierarchical classification (AHC) | 66 |
| Geographical descrimination of honeys from North and central Mozambique | Elecrical conductivity, moisture, 3-methylbutan-1-ol and free acidity | P&T-GC/MS + sugars + physicochemical parameters | ANOVA, PCA (2 PCs explained 67% of the variance), SLDA (correct classification of 96.7%) | 80 |
| Discrimination of honey Honey collected by Apiscerana and Apis mellifera | Benzaldehyde, heptanal, phenylacetaldehyde, trans-linalool oxide, 1-nonanol, phenethyl acetate, 1-heptanol, cyclohexanone | HS-GC-IMS + HS-SPME-GC-MS | PCA (2 PCs explained 62.3% of the variance), OPLS-DA, VIP analysis | 72 |
| Botanical discrimination of Greek unifloral honeys (citrus, fir, pine, and thyme) | Dill ether, alpha-4-dimethyl-3-cyclohexene-1- acetaldehyde, acetic acid ethyl ester, octanoic acid ethyl ester, methylanthranilate, 2,2,4,6,6-pentamethyl-heptane, phenylacetaldehyde, cis-linalool oxide, lilac aldehyde (isomer III) | HS-SPME-GC/MS | MANOVA, LDA (correct classification of 84.1%), SLDA (3 DF explained 100% of total variance), (correct classification of 93.9%), kNN (correct classification 89.5%) | 64 |
GC, coupled with single or triple quadrupole mass analyzers (GC-MS), is the most utilized analytical technique for volatomic studies. LLE has been used, but in most cases, HS-SPME is the choice to extract phenolic compounds since it is simple, fast, sensitive and versatile, as different fiber coatings are available. Fibers containing more than one coating have been most commonly employed due to their capability to retain a broader range of compounds. HS-SPME-GC-MS using divinylbenzene – carboxen-poly (dimethylsiloxane) (DVB/CAR/PDMS) has been successfully applied in combination with chemometric tools to discriminate Greek unifloral honeys, as well as to geographically differentiate pine and thyme honeys from different regions in Greece.59–61 The capability of this technique has been broadly cited in the literature. Thus, Silva et al. managed to differentiate certified and non-certified sugarcane honey,62 Madas et al. discriminated acacia honey samples from different regions in Romania,63 and Karabagias et al. verified the floral source of Greek unifloral honeys.64 Other multi-coating fibers used in volatomic studies are PDMS/DVB and PDMS/CAR. Costa et al.65 distinguished honeys derived from different botanical and entomological origins, while Neggad et al.66 geographically differentiated Algerian honeys from Arid and Mediterranean Areas. Headspace analysis can be conducted in either a static or a dynamic way. Static HS-GC-MS is a powerful option for volatile compound extraction, as it is relatively cheap, straightforward and can be easily automated, but it may be less sensitive than the other techniques. Eleven volatile compounds were found to differentiate honey from different provinces in Brazil produced by 8 species of stingless bees after PCA and LDA chemometric analysis.67 Purge and trap (P&T) is a type of dynamic HS, and is more sensitive than static HS. It is preferred for the analysis of semi and higher boiling volatiles, but cannot normally be automated. Many researchers have successfully applied the P&T-GC-MS technique combined with other parameters, such as physicochemical parameters and sugars, to assess honey authenticity. Juan-Borrás et al.68 differentiated acacia, sunflower and tilia honey from three different countries, Escriche et al.69 verified the label description of Levanter and Thyme honeys, and Tanleque-Alberto et al.70 discriminated honeys from North and central Mozambique.
Besides GC-MS, other analytical platforms have been utilized for honey authenticity using the volatile fraction. GC-ion mobility Spectrometry (GC-IMS) combined with headspace sampling (HS-GC-IMS) has been proved as an efficient separation method for honey authenticity studies. Its sensitivity, simplicity, robustness and low-cost led to the employment of this technique for the botanical discrimination of European acacia, canola and honeydew honeys.71 The collected data were statistically processed using PCA, LDA and k-nearest neighbors (k-NN), and reveals that hexanal, cis-linalool oxide, benzaldehyde, 3-hydroxy-2-butanone, trans-2-pentenal, and 3-methylbutanol are markers for European honeys. HS-GC-IMS profiling, in conjunction with statistical tools, has also been applied to differentiate honey produced by Apis cerana and Apis mellifera bees.72 Additionally, HS-SPME-GC-Ion Trap using a DVB/CAR/PDMS fiber has been used to study orange honey authenticity, revealing that linalool and linalool oxide isomers are markers for this type of honey.73 Last but not least, HRMS offers high mass accuracy and resolution, providing selectivity and a powerful tool to identify unknown compounds due to the wealth of MS and MS/MS data acquired. The combination of HRMS, such as TOF with HS-SPME-GC, with a chemometric workflow can serve as an effective approach in volatomic studies. Toluene has been proposed to discriminate mono and multi-flora honeys from three countries using HS-SPME-GC-QTOF-MS and PCA for dimension reduction.74 Moreover, benzoic acid, isophorone and 2-methylbutyric acid, among others, were also proposed as markers for the discrimination of the floral origin of heather, raspberry, rape and alder buckthorn honeys utilizing HS-SPME-GC-TOF.75
Apart from volatile compounds, GC techniques have been employed to determine other compounds, such as amino acids and sugars. Such analytes require a derivatization step before gas chromatographic analysis to change their properties for better separation and sensitivity. As these compounds are not volatile, liquid injection is used to transfer analytes in a gas chromatographic system. Azevedo et al. determined the free amino acid profile, and managed to discriminate bracatinga honeydew honeys from different regions using GC-MS and multivariate statistical principal component analysis.76 Finally, Pascual-Mate et al. distinguished the botanical origin of honeydew, multifloral, chestnut, heather, lavender and clover honeys from the northern Iberian plateau, determining the sugar composition using GC coupled to a flame ionization detector (FID), the moisture content and the specific rotation.77
3.3 Bioanalytical techniques
Techniques targeting genetic material have been thoroughly used in the food authentication field. The genetic data coded in the DNA can provide the necessary information about the botanical, geographical and/or genetic origin. Polymerase Chain Reaction (PCR) is a sensitive technique used for the amplification of specific DNA parts, starting from a small sample or tissue quantity.81 In honey authenticity studies, DNA-based methods have been used in order to determine the botanical and entomological origin of honey samples (Table 4). For the botanical origin investigation, plastid genome regions have been used as biomarkers. I. Bruni et al.82 targeted the rbcL and trnH-psbA plastid regions of 4 multifloral honey samples using a reference DNA barcoding database and PCR. The results obtained led to over 99% DNA match for all the flower species (12, 14, 14 and 15 plant species). A PCR method accurately detected mono- and multifloral honey samples by targeting the cyt2b, matk, psbA, and ndhF genes of plant pollen.83 After DNA extraction, real-time PCR was used for the amplification of the selected genes. The results showed that from the 159 samples (all characterized as monofloral), 11 samples (7%) were found to also contain pollen from other plants, and were thus mislabeled as monofloral. In another paper,84 the botanical authentication of lavender (Lavandula spp.) honey samples was achieved by targeting the plastidial matK gene of the Lavandula species contained in the honey samples. Endpoint and real-time PCR were used for gene amplification. The results showed three distinct clusters (different lavender species) with high level of confidence (>99%). Regarding the entomological origin of honeys,85 a methodology targeting the ribosomal RNA (16S rRNA) gene region and mitochondrial cytochrome c oxidase subunit I (COI) gene region of the bee DNA was proposed. The gene markers 287 bp 16S rRNA gene and 284 bp COI gene were used to classify and correctly differentiate the honey samples produced by Apis honey bees and Trigona stingless bees. Sónia Soares et al.86 proposed a new method for the authentication of European and Asian honey samples by targeting the tRNAleu-cox2 intergenic region and the 16S rRNA gene of both European and Asian bee species. The developed methodology correctly identified 9 Asian honeybee and 6 European honeybee samples.| Authenticity issue | Markers | Method | Chemometric tool | Ref. |
|---|---|---|---|---|
| Botanical composition investigation of 4 multifloral honeys | DNA barcoding of rbcL and trnH-psbA plastid regions | DNA analysis (PCR) | >99% DNA match for every flower species | 82 |
| Botanical origin identification of 3 monofloral and one multifloral honey | PCR primers were used to detect adh1 gene of heather (C. vulgaris) | DNA analysis (PCR) | Adh1 gene of heather (originating from Portugal) was found in all samples | 87 |
| Identification of honey entomological origin (5 unifloral honeys) | The 300 bp of mitochondrial large subunit ribosomal RNA (16S rRNA) gene region and mitochondrial cytochrome c oxidase subunit I (COI) gene region | PCR amplification | Correctly classify and differentiate honey samples based on entomological origin | 85 |
| DNA sequencing | ||||
| Molecular tracing of the botanical origin of honey samples | cyt2b, matk, psbA, and ndhF genes | Real-time PCR | Method accurately detected mono- and multifloral honey | 83 |
| Entomological origin identification of honey | Mitochondrial 16S rRNA gene | PCR amplification, DNA sequencing and BLAST analysis for species identification | One-way ANOVA | 88 |
| Asian (Apis cerana) and European (Apis mellifera) honey entomological authentication | mtDNA region located between the tRNAleu and cytochrome c oxidase subunit II genes tRNAleu-cox2 intergenic region and 16S rRNA | Real-time PCR with high resolution melting (HRM) | Correct identification of samples entomological origin | 86 |
| Botanical authentication of lavender (Lavandula spp.) honey | Plastidial matK gene | DNA-barcoding coupled to high resolution melting analysis (HRM) & end-point PCR | 99% confidence of three clusters: Portugal lavender species, the species L. multifida & L. pinnata and French lavender species | 84 |
| Entomological origin investigation applied to Sicilian honey bee (A. m. siciliana) and Iberian honey bee (A. m. iberiensis) honeys | mtDNA haplotype variability | PCR | Correct discrimination of three honey branches | 89 |
3.4 Spectroscopy
Chemical information gathered by spectroscopic techniques can be a valuable asset towards honey authentication (Table 5). A huge advantage offered by such techniques is the ability to record spectral information quickly and effectively, without the need of complex sample preparation protocols.90 Furthermore, the majority of these methods rely on the fingerprint profiling of honey samples, and not on the determination of selected analytes. This can facilitate the analytical procedure by minimizing the analysis and data treatment time. Spectral data can provide information on the honey chemical profile, and the application of chemometric tools is necessary for differentiation and discrimination. The botanical origin of Estonian honey was investigated by using front face fluorescent spectroscopy assisted by PARAFAC algorithm.91 Spectral fluorescence signatures were acquired at excitation wavelengths from 230–350 nm, and at emission wavelengths from 250–565 nm. The PARAFAC algorithm was applied to the raw spectral data, distinguishing the raspberry honey samples from the others. Polyflolar and monofloral honey samples were classified by using front-face synchronous fluorescence spectroscopy combined with PCA and PLS.92 The developed model attained 88.3% successful prediction of polyfloral honey, while three PCs explained more than 95% of the variance between samples.| Authenticity issue | Markers | Method | Chemometric tool | Ref. |
|---|---|---|---|---|
| Estonian honeys botanical origin characterization | Spectral fluorescence signatures | Front-face fluorescence spectroscopy | PARAFAC algorithm, PCA Correlation (r2) between experimental data and estimated values was higher than 0.8 | 91 |
| Classify honey samples according to their botanical origin and distinguish fake from natural honey | Spectral fluorescence spectra in an excitation range of 240–500 nm for synchronous wavelength intervals of 30–300 nm | Front-face synchronous fluorescence spectroscopy | PCA, PLS-DA 88.3% successful prediction of polyfloral honey | 92 |
| Artificial honeys well separated from natural honey | ||||
| Fluorescence characteristics of New Zealand honeys examination | Two excitation–emission (ex–em) marker wavelengths each for manuka and kanuka honeys (MM1 & MM2) | Fluorescence spectroscopy | Northland, Waikato Wetlands, and East Coast manuka honeys showed significant differences at MM2 | 100 |
| Botanical origin classification and adulterant determination of raw honey. | NIR & MIR spectra of honey samples | NIR & MIR spectroscopy | PLS-DA accuracy for calibration and prediction sets >96% | 93 |
| Investigation of authenticity and fraud detection in South African honey | NIR spectra of honey samples | NIR spectroscopy | PLS-DA classification accuracies >93.3% | 101 |
| Identification and classification of honey's authenticity (entomological origin) | ATR-FTIR spectra of honey samples | ATR-FTIR spectroscopy | Discriminant analysis, performance Index >87.7% | 94 |
| Wavelengths that can best differentiate: 1600–1700 cm−1; 1175–1540 cm−1; 940–1175 cm−1; and 700–940 cm−1 | ||||
| Organic and conventional differentiation of Italian honey samples | Succinate and acetate for conventional, kynurate for organic | 1H NMR | PCA, PLS-DA, Q2 > 65% for polyfloral and >98.1% for unifloral honey samples | 102 |
| Origin and composition investigation of European acacia honeys based on geographical floral markers | NMR fingerprinting | 1H NMR | PLS2-DA, 100% correct classification rate | 98 |
| Acacia honey authenticity | Profile of 20 minor saccharides | 1H NMR | PCA, PC1 + PC2 explain 81% of the total variance | 99 |
IR spectroscopy can also provide reliable results when the botanical and/or entomological origin of honeys is the scope of the research. Both near-infrared (NIR) and mid-infrared (MIR) regions of the electromagnetic spectrum have been used for the detection of the botanical and entomological origin of honey samples. Zhilin Gan et al.93 acquired the NIR and MIR spectra of raw honey samples. With the help of PLS-DA and PCA, they managed to classify honeys of different botanical origin with accuracy and prediction at greater than 96%. Sahlan et al. investigated the entomological origin of 58 honey samples (produced by 5 bee subspecies, 3 from Apis spp and 2 of Tetragonula spp) by attenuated total reflectance Fourier-transform infrared spectroscopy (ATR-FTIR) coupled with DA.94 The wavelengths that best classified the two species is the region between 1600–1700 cm−1.
NMR spectroscopy has been a very reliable technique used by analytical scientists in the food authenticity field for over two decades,95 mostly because of its robustness, its ability to detect compounds in complicated mixtures without prior purification and/or separation, and the simple-to no sample preparation. NMR spectroscopy has been applied in honey authenticity studies with very satisfying results, in combination with multivariate chemometric tools. The great advantage of NMR analysis is that no individual compounds need to be identified. However, a fingerprint profile of each sample is sufficient in order to determine or predict the sample botanical, geographical or entomological origin. It is important to emphasize that chemometric tools are necessary to extract useful conclusions, considering the complexity of the NMR spectra.
More than 800 honey samples of mono- and multifloral origin from various geographic regions were analyzed using 1H-NMR.96 Both targeted and non-targeted approaches have been followed in this study. For the targeted compounds, typical honey quality markers were identified and quantified (glucose, fructose, sucrose, 5-HMF). For the untargeted approach, the whole spectrum was processed. Independent Components Analysis (ICA) was used for the extraction of analytical important signals by discriminating the spectral data originating from the source molecule. The results showed specific markers for some of the monofloral honeys, providing a reliable solution for the botanical origin determination. Metabolic fingerprint of both water and chloroform honey extracts were used22 to determine the entomological origin of honeys. Seventy-eight genuine Ecuadorian honeys originating from 4 bee species (23 from Apis mellifera, 16 from Geotrigona-Trigona, 15 from Melipona and 24 from Scaptotrigona) were analyzed with minimum sample preparation with 1H-NMR. The orthogonal projections to latent structures-discriminant analysis (OPLS-DA), correctly classified from 87% to 100% of the samples by using the water extracts. The spectra acquired from the chloroform extracts presented significant differences between the different species. These differences occurred due to the extracted waxes or cerumen pots. In this way, the entomological related marker compounds can be identified correctly. Chloroform honey extracts were also utilized to acquire the 1H-NMR profile of 983 samples originating from 16 plant species.97 The acquired data were used in order to build an OPLS-DA model, in which not only the monofloral species were identified, but also the polyfloral samples. For the majority of the analyzed samples, the sensitivity was above 90%, resulting in a reliable method for investigating the botanical origin of honey. In another study published by the group,98 217 honey samples, declared as acacia honeys, were obtained from local markets in Italy. The 1H-NMR profiling of the chloroform extracts classified all honey samples as monofloral acacia honeys, assisted by using PCA and PLS-DA. Moreover, a clear discrimination based on the geographical origin was achieved. Italian and Eastern European honeys were differentiated due to the differences presented in the secondary metabolite composition. Furthermore, the proposed methodology revealed the capability of characterizing the blends and mixtures of Italian and EU originating honeys. Importantly, the sugar composition of honey samples can also provide information about their geographical origin. A simple dilution of the sample in a phosphate buffer solution, followed by 1H-NMR analysis, can provide the necessary information for the sugar composition of honey.99 In this study, Chinese (n = 16 samples) and European honeys (n = 46 samples) presented significant differences in their sugar composition. Chinese honeys featured lower content of minor disaccharides, while higher levels of monosaccharides (glucose, fructose and mannose) were reported. European honeys did not have any significant differences in their sugar content among them. Using PCA, two clusters have been formed (one of the EU and one of the Chinese samples), with two PCs accounting for the 81% of the total variance. Furthermore, this method can give important information about honey adulteration using industrial syrups for the feeding of bee colonies during the main nectar flow period by determining some sugar molecules (e.g., mannose) that are not normally present in genuine acacia honeys.
3.5 Elemental techniques
The honey elemental content is affected by various factors, such as geographical origin, botanical origin, environmental conditions and anthropogenic activity.103 In detail, the minerals in honey mainly come from nectar or honeydew and pollen grains.104,105 Concerning their geographical origin, the metal content of honey is mainly derived from soil. Metals are transported from the soil to honey plants through the root system, pass on to the nectar, and then to the honey produced by bees. Soil composition is determined by geochemical and geological features, such as hydrothermal and possible volcanic activity or climatic conditions in the forage area of the bees. As honey reflects the elements of the plants from which the honeybees collect their food, the elemental fingerprint of honey depends on the type of soil, as well as the type of plant.106,107 Therefore, the flower elemental composition plays a decisive role, and is also related to the honey botanical origin.106 The floral type of honey plants, floral density, and the chemical composition of nectar, pollen, and other forage sources significantly differ depending on the location of apiaries and the vegetation type.108 Vegetation season is also important as different elements have different concentrations in honey from the same botanical type even when collected from the same geographical region, same locality, and same beehive.109 Environmental factors and climatic conditions also affect the elemental content.110 The metal presence in honey can also be due to anthropogenic activities, e.g., industrial and agricultural practices and landfills.111 As shown in Fig. 6, elemental techniques are most commonly used in geographical origin studies, but also in other authenticity cases.The elemental fingerprint is predominantly determined through inductively coupled plasma mass spectrometry (ICP-MS) and inductively coupled plasma optical emission spectroscopy (ICP-OES), as they provide multielemental determination within a single run (Table 6). Regarding sample preparation, closed-vessel microwave-assisted digestion is the common practice.103 Elemental composition can be used as an indication of the botanical and geographical origin of honey. In a recent study, Squadrone et al.106 determined 40 elements, including rare earth elements (REEs), by ICP-MS in 91 monofloral and multifloral honeys from Piedmont, Northwestern Italy. The differences in the trace element concentrations between the honeys of different botanical origin were observed, while REEs could help in discriminating monofloral and multifloral honeys via the light rare earth element to heavy rare earth element (LREE/HREE) ratio. In order to discriminate honey according to the various floral types, PCA and Analysis Of Variance (ANOVA) were used. PCA revealed that the metal content could provide enough information to develop a first classification. Another recent study found significant differences in the trace and rare earth element content in honeys from different countries, especially Tanzanian honeys.112 Rare earth elements occur in rocks and are then translocated by diverse natural processes, like biological activity and weathering. Subsequently, the REEs content is minimally affected by the harvesting year, indicating their potential for geographical origin discriminations. According to Magdas et al., REEs are considered more efficient for the discrimination of unprocessed food matrices (e.g., vegetables, fruits, honey).113–115 Lanthanum, Ce, Pr, Nd, Sm, Eu and Gd. (LREEs) present higher mobility in plants. For this reason, they present higher potential for geographical origin discrimination in comparison to the rest of the REEs.113,116 Manganese and Pr & La are correlated with the honey varieties distinction.113
| Authenticity issue | Markers | Method | Chemometric tool | Ref. |
|---|---|---|---|---|
| Botanical origin | Fe, Mn, Zn, Cu and Hg | ICP-MS | ANOVA, PCA, LDA | 121 |
| Acacia, sunflower, tilia, and polyfloral from 3 Romanian regions | 80% successful botanical origin discrimination using LDA | |||
| Botanical origin | Na, Mg, K, Ca, Mn, Fe, Cu, Rb, Sr, Ba | ICP-MS | PCA, PLS-DA, BP-ANN | 122 |
| Inden, vitex, rape, and acacia, collected from 4 Chinese regions | 100% accuracy for linden, vitex, and rape honey samples | |||
| 92.3% accuracy for acacia honey and rape honey | ||||
| Botanical origin | Al, Ca, Cu, Fe, Mg, Mn, Sb, Si and Zn | ICP-OES | MANOVA, LDA, k-NN and MCA | 123 |
| 270 citrus, fir, multifloral, pine and thyme from Greece, Cyprus, Egypt, Spain, and Morocco | ||||
| Botanical origin | K, Ca, Mg, Na, P and S | ICP-OES | One-way ANOVA and LDA | 124 |
| 140 Hungarian mono-floral honey samples (acacia, linden, sunflower, rape, chestnut, forest, silk grass, and facelia) | 96% botanical origin prediction using LDA | |||
| 100% botanical origin prediction using K/Na and K/Mg ratios | ||||
| Geographical origin | Ag, Al,As, B, Ba, Be, Cd, Co, Cr, Cu, Fe, Mg, Mn, Mo, Ni, Pb, Sb,Se, Sn, and Zn | ICP-OES | PCA and LDA | 125 |
| North, west, east, and south regions of Johor, Malaysia | Cross-validation using PCA demonstrated 87.0% correct classification rate, while 96.2% with the use of LDA | |||
| Geographical origin | Ag, Al, As, B, Ba, Be, Ca, Cd, Co, Cr, Cu, Fe, Hg, Mg, Mn, Mo, Ni, Pb, Sb, Se, Si, Ti, Tl, V, and Zn | ICP-OES | MANOVA and LDA | 126 |
| 39 pine and 42 honey samples from 9 different regions of Greece | The correct prediction rates were 84.6 and 83.3% for pine and thyme honeys, respectively | |||
| Geographical origin and time-dependent composition | Na, Ca, Mg, K, Al, B, Ba, Bi, Cd, Co, Cr, Cu, Fe, Li, Mn, Ni, Pb, Sr and Zn | MP-AES | One-way ANOVA and CDA | 119 |
| Acacia samples from 3 Hungarian region collected from 1958–2018 | ||||
| Geographical origin | Ag, Al, As, Au, B, Ba, Be, Bi, Ca, Cd, Ce, Cs, Cr, Co, Cu, Dy, Er, Eu, Fe, Ga, Gd, Ge, Hg, Hf, Ho, Rb, K, La, Li, Lu, Mg, Mn, Mo, Na, Nb, Nd, Ni, Os, P, Pb, Pd, Pt, Pr, Re, Ru, Se, Sb, Sr, Sm, Sn, Ta, Tb, Te, Th, Tl, Tm, Ti, U, V, W, Y, Yb, Zn and Zr and δ13Cprotein (‰) | ICP-MS, IRMS | ANOVA, PCA and CDA, PCA and CDA coupled with C5.0 classification modelling of honey carbon | 127 |
| Commercial honey samples from 5 different continents | Trace elements from Australian regions differ statistical from other continents. | Isotopes and trace elements showed distinct clusters according to their geographic origin | ||
| The C5.0 model revealed that Sr, P, Mn and K can be used to differentiate geographic origin |
There have also been some novel approaches, which eliminate the need for sample preparation, namely laser-induced breakdown spectroscopy (LIBS)117 and energy-dispersive X-ray fluorescence (ED-XRF).118 Zhao et al.117 used LIBS to rapidly trace the geographical origin of acacia and multi-floral honeys. Magnesium, Ca, Na, and K LIBS emissions presented significant differences among different geographical origins. Principal Component Analysis, Support Vector Machines (SVM) and LDA were used as the chemometric techniques. Support Vector Machines performed better than LDA, while discriminant results were better for multi-floral than acacia honey. Some deep learning methods, such as convolutional neural networks, might be used to further improve the performance. Macro- and trace elements were determined by ED-XRF, and multivariate analysis was utilized to classify honey according to the botanical variety and geographical origin.118 For the creation of a classification model, PCA and PLS-DA were used to classify 9 botanical types (orange, robinia, lavender, rosemary, thyme, lime, chestnut, eucalyptus and manuka) and 7 geographical origins, namely Italy, Romania, Spain, Portugal, France, Hungary and New Zealand. Furthermore, PLS-DA models for the specific combinations of botanical variety-country allowed for the successful classification of samples, and were verified by external validation samples. Although the number of different botanical types and geographical origins was high, this was not the case for the number of samples. A microwave plasma atomic emission spectrometer (MP-AES) was also used for the elemental analysis of Hungarian honey samples, as it could be a cost-effective alternative for metal content determination.119 It is worthwhile to notice that this study exploited the elemental content of honey as an environmental change indicator related to natural and anthropogenic causes (samples collected from 1958 to 2018). Besides the aforementioned methods, the implementation of high-resolution ICP-MS (HR-ICP-MS) has introduced novel analytical opportunities, which resulted in the so-called “elemental metabolomics”. Elemental metabolomics have the potential to assist honey authenticity through its thorough elemental characterization.120 Up to 270 elemental isotopes can be measured using HR-ICP-MS, which is a great advancement in comparison to the 70 elements that may be measured using conventional ICP-MS.
3.6 Isotopic techniques
Stable isotope ratios are influenced by the climatic conditions, geographical origin and geological factors. Hydrogen and oxygen isotopes originating in organic matter are connected to the H and O isotope data of water. Nitrogen and carbon isotopes are associated with climatic conditions and agricultural practices.128 The isotopic fingerprint has been used in honey origin studies (Table 7). Bontempo et al.129 studied Italian honeys of different botanical origin (polyfloral, citrus, rhododendron, eucalyptus, acacia, chestnut and honeydew). PCA was used and the first component was mainly negatively loaded by the δ13C of honey, ethanol and proteins, and positively by K, Mg, Ca, Rb and Ba. In contrast, the second was mainly negatively loaded by δ15N, B, Na and (D/H)I. Regarding the geographical origin, only chestnut honeys were used without a clear separation of all of the regions. The first component was positively loaded by the δ13C of honey, ethanol and proteins, and negatively by Rb, Sr and B. In contrast, the second principal component was mainly positively loaded by δ15N proteins and negatively by Mn. Higher δ15N could be associated with the fact that citrus and eucalyptus plants normally grow closer to the sea, while lower δ15N values of rhododendron honeys could be associated with the higher altitudes where this plant normally grows. Rhododendron honeys presented low concentrations of B and Ca because rhododendron plants usually prefer acidic soils.| Authenticity issue | Markers | Technique/method | Chemometric tool | Ref. |
|---|---|---|---|---|
| Geographical and botanical origin | δ13C, δ18O and δ2H along with (D/H)I from ethanol | IRMS and sitespecific natural isotopic fractionation (SNIF) - NMR | PCA | 133 |
| Acacia (Robinia pseudoacacia), sunflower (Helianthus annus), linden (Tilia platyphyllos), rape (Brassica napus oleifera), honeydew, and multifloral honey samples from Romania | ||||
| Commercial samples from Italy, Russia, and Turkey | ||||
| Botanical and geographical origin | δ13C of honey, ethanol and proteins, (D/H)I, δ15N of protein Al, B, Ba, Ca, Cr, Cu, Fe, K, Mg, Mn, Na, Ni, Pb, Rb, Sr, Zn | IRMS, SNIF-NMR and ICP-OES | PCA | 129 |
| Different botanical origin (polyfloral, citrus, rhododendron, eucalyptus, acacia, chestnut and honeydew) produced throughout Italy in different years | δ13C of honey, ethanol and proteins, δ15N, (D/H)I, K, Mg, Ca, Rb, Ba, B and Na for botanical origin & δ13C of honey, ethanol and proteins, δ15N, Rb, Sr, B and Mn for geographical origin | |||
| Botanical origin | δ13C, colour intensity, radical scavenging activity, P and Sn | IRMS and ICP-OES | MANOVA and LDA | 134 |
| 4 botanical species (thyme, pine, fir, orange blossom) from 4 Greek regions | ||||
| Geographical origin | δ13C value, oligosaccharides and polyphenols | IRMS, GC-MS | PCA and PLS-DA | 132 |
| Acacia honey from 6 different Chinese regions | Lower δ13C of honey from Gansu than those of other regions | HPLC-MS | ||
| Higher oligosaccharides from Shanxi and Shaanxi regions than other four regions. Polyphenols from Shandong was the highest and were better parameters than both δ13C and oligosaccharides for geographical origins discrimination | ||||
| PLS-DA showed that when all 31 different parameters were combined, a classification rate of 94.12%. | ||||
| Could be achieved using external cross validation method | ||||
| Botanical origin | d13C data of honey, protein fraction, and the isotopic index | IRMS | Logistic regression model | 135 |
| Eucalyptus and pasture honey from Uruguay | ||||
| Botanical origin | Physicochemical properties, major sugar composition and δ13C signature of honeys | Physicochemical analysis, HPLC-ELSD, IRMS | ANOVA, HCA and PCA | 136 |
| Acacia (Robinia pseudoacacia) rape (Brassica napus oleifera), sunflower (Helianthus annus), linden (Tilia platyphyllos) polyfloral, honeydew samples | δ13C values of protein extracted from honey, glucose content, ratio between Fructose and glucose, and electrical conductivity were significantly different, depending on the botanical origin of honeys |
In a very recent study, Magdas et al.113 examined emerging markers for geographical and varietal discrimination of Romanian and French honey. The isotopic fingerprint of honey water, carbon and hydrogen isotopic ratios and REEs were used as emerging markers. Results showed a geographical differentiation higher than 98%, while the markers (D/H)I, δ2H, δ18O, La, Ce and Pr especially played an essential role. Floral recognition presented a lower percentage, showing that these markers are more suitable for geographical classification. The observed differences of δ2H and δ18O between the French and Romanian honeys are attributed to the different geo-climatic conditions.
In addition, honeydew honey authenticity was studied. Vasić et al.130 tried to discriminate five varieties of honey. They determined SCIRA, 20 elements and 14 carbohydrates. Several elements (Ba, Ca, Mg, Sr, Mn, Al, Co, Ni, Se) were indicated as characteristic of the honey type, and allow for the classification of three botanical origins (Abies alba, Quercus frainetto, Quercus ilex). However, none of the sugar compounds were exploited as a marker. However, the sugars turanose, trehalose, arabinose and raffinose, elements Ba, Sr, P, Cd and Se, and δ13C values of honey showed significant differences, according to the production year. Importantly, besides an adulteration indicator, the protein δ13C could also be a botanical origin indicator. In addition, honeydew honeys showed a higher amount of mineral compounds in comparison to blossom honey. In the same study, Hungarian oak honeydew honey was clearly discriminated from other honeys due to the alkaline earth elements, the Mn and S high content and the high δ13C values of protein. Overall, the low number of the 2 studied honey varieties rendered the results as preliminary. In another interesting study, hydrogen, carbon, nitrogen and sulphur stable isotope ratios of honey protein were determined in 516 honey samples from 20 European regions. The authors stated that the mean hydrogen isotopic ratios were associated with the mean hydrogen isotopic ratios of precipitation and groundwater of the regions, while the carbon isotopic ratios were affected by climate, sulphur stable isotope composition and the surface geology. The isotopic profile of these elements could provide useful origin information.131 Last but not least, isotopic analysis was combined with oligosaccharide and polyphenol content to discriminate the geographical origin of Chinese honey.132 The IRMS data were fused with GC-MS and HPLC-MS analytical information, achieving a 94% correct classification by using PLS-DA. This is a striking example of the analytical technique combination to increase the model classification power in food authenticity studies.
4. Conclusions
Analytical techniques to verify the honey origin have drawn ever-increasing attention, indicating that it is an emerging trend in the food authenticity field. Research on honey authenticity is connected to certain inherent difficulties in comparison to other food commodities. In detail, fruits and vegetables are grown in a well-defined area, while breeding animals have a controlled diet. On the contrary, bee forage area could extend up to more than 5 km (radius) form their hive,113 showing that contradictory results among studies may be related to this fact (for example, in the case of using elemental techniques to define honey origin). The reviewed techniques were used to investigate different concepts. Chromatographic separation combined with various detectors was mostly used to determine the honey botanical source. Similarly, physicochemical honey characteristics (such as free acidity, color or electrical conductivity) were also mostly used in botanical origin studies. Actually, the honey physicochemical characteristic determination was mostly initially applied, as it requires minimal instrumentation and the cost of such analysis is low in comparison to instrumental techniques. In terms of the molecular techniques, these were mostly used to determine the floral and entomological honey source, as they use the specified DNA parts as markers, which are characteristic for specific species. Spectroscopic techniques provided the necessary information to assess the botanical, geographical or entomological origin requiring minimal sample preparation. In any case, a high number and a variety of samples are necessary to build reliable spectral databases/libraries. Elemental profiles proved to be reliable indicators for geographical origin determination in many cases, while more work is needed to investigate the relation to genetic origin. Isotopic techniques are usually combined with other techniques, for example elemental or chromatographic; an approach that attained promising results in geographical and botanical origin determination. Besides botanical and geographical origin, novel authenticity issues emerged, namely, production type, discrimination between honeydew and blossom honey, and honey entomological source, which will be studied in the future in our view. All in all, the interest in the development of analytical methods for honey origin determination is expected to grow, enhancing food quality and assuring product origin.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge support of this work by the project ≪FoodOmicsGR Comprehensive Characterisation of Foods≫ (MIS 5029057), which is implemented under the Action “Reinforcement of the Research and Innovation Infrastructure”, funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).References
- D. Cianciosi, T. Forbes-Hernández, S. Afrin, M. Gasparrini, P. Reboredo-Rodriguez, P. Manna, J. Zhang, L. Bravo Lamas, S. Martinez Florez and P. Agudo Toyos, Molecules, 2018, 23, 2322 CrossRef PubMed.
- M. G. Miguel, M. D. Antunes and M. L. Faleiro, Integr. Med. Insights, 2017, 12, 1–15 CrossRef PubMed.
- P. M. da Silva, C. Gauche, L. V. Gonzaga, A. C. O. Costa and R. Fett, Food Chem., 2016, 196, 309–323 CrossRef PubMed.
- D. Cianciosi, T. Y. Forbes-Hernández, J. Ansary, E. Gil, A. Amici, S. Bompadre, J. Simal-Gandara, F. Giampieri and M. Battino, Food Chem., 2020, 325, 126881 CrossRef CAS PubMed.
- S. Afrin, S. M. Haneefa, M. J. Fernandez-Cabezudo, F. Giampieri, B. K. Al-Ramadi and M. Battino, Nutr. Res. Rev., 2020, 33, 50–76 CrossRef CAS PubMed.
- Y. Ranneh, A. M. Akim, H. A. Hamid, H. Khazaai, A. Fadel, Z. A. Zakaria, M. Albujja and M. F. A. Bakar, BMC Complementary Med. Ther., 2021, 21, 30 CrossRef PubMed.
- N. L. García, Bee World, 2018, 95, 89–94 CrossRef.
- A. Hassoun, I. Måge, W. F. Schmidt, H. T. Temiz, L. Li, H.-Y. Kim, H. Nilsen, A. Biancolillo, A. Aït-Kaddour and M. Sikorski, Foods, 2020, 9, 1069 CrossRef CAS PubMed.
- S. Bogdanov and P. Martin, Swiss Bee Research Centre, 2002 Search PubMed.
- A. J. Siddiqui, S. G. Musharraf, M. I. Choudhary and A. Rahman, Food Chem., 2017, 217, 687–698 CrossRef CAS PubMed.
- M. Yang, Y. Gao, Y. Liu, X. Fan, K. Zhao and S. Zhao, J. Agric. Sci. Technol., 2018, 20, 685–693 Search PubMed.
- L. Wu, B. Du, Y. Vander Heyden, L. Chen, L. Zhao, M. Wang and X. Xue, TrAC, Trends Anal. Chem., 2017, 86, 25–38 CrossRef CAS.
- K. W. Se, R. A. Wahab, S. N. Syed Yaacob and S. K. Ghoshal, J. Food Compos. Anal., 2019, 80, 16–32 CrossRef CAS.
- C. Pita-Calvo and M. Vázquez, Trends Food Sci. Technol., 2017, 59, 79–87 CrossRef CAS.
- I. Jasicka-Misiak, E. Makowicz and N. Stanek, Eur. Food Res. Technol., 2018, 244, 1169–1184 CrossRef CAS.
- B. Khakimov, G. Gürdeniz and S. B. Engelsen, Acta Aliment., 2015, 44, 4–31 CrossRef CAS.
- D. Granato, P. Putnik, D. B. Kovačević, J. S. Santos, V. Calado, R. S. Rocha, A. G. Da Cruz, B. Jarvis, O. Y. Rodionova and A. Pomerantsev, Compr. Rev. Food Sci. Food Saf., 2018, 17, 663–677 CrossRef PubMed.
- C. Maione, F. Barbosa and R. M. Barbosa, Comput. Electron. Agric., 2019, 157, 436–446 CrossRef.
- S. Soares, J. S. Amaral, M. B. P. P. Oliveira and I. Mafra, Compr. Rev. Food Sci. Food Saf., 2017, 16, 1072–1100 CrossRef CAS PubMed.
- V. Berriel, Foods, 2018, 7, 86 CrossRef PubMed.
- G. Bergamo, S. K. T. Seraglio, L. V. Gonzaga, R. Fett and A. C. O. Costa, Food Res. Int., 2019, 116, 745–754 CrossRef CAS PubMed.
- V. Zuccato, C. Finotello, I. Menegazzo, G. Peccolo and E. Schievano, Food Control, 2017, 82, 145–153 CrossRef CAS.
- E. Guzelmeric, I. Ciftci, P. I. Yuksel and E. Yesilada, LWT, 2020, 132, 109921 CrossRef CAS.
- R. Balkanska, K. Stefanova and R. Stoikova–Grigorova, J. Apic. Res., 2020, 59, 852–861 CrossRef.
- S. Bogdanov, P. Martin and C. Lullmann, Harmonised methods of the international honey commission, Swiss Bee Res Centre, FAM, Liebefeld Search PubMed.
- D. C. Fechner, A. L. Moresi, J. D. Ruiz Díaz, R. G. Pellerano and F. A. Vazquez, Food Biosci., 2016, 15, 49–54 CrossRef CAS.
- S. Popek, M. Halagarda and K. Kursa, LWT, 2017, 77, 482–487 CrossRef CAS.
- I. K. Karabagias, M. V Vavoura, C. Nikolaou, A. V Badeka, S. Kontakos and M. G. Kontominas, Food Res. Int., 2014, 62, 753–760 CrossRef CAS.
- P. Combarros-Fuertes, R. M. Valencia-Barrera, L. M. Estevinho, L. G. Dias, J. M. Castro, M. E. Tornadijo and J. M. Fresno, J. Apic. Res., 2019, 58, 92–103 CrossRef.
- I. Rodríguez, S. Serrano, H. Galán-Soldevilla, G. Piva and J. L. Ubera, Int. J. Food Sci. Technol., 2015, 50, 1545–1551 CrossRef.
- M. E. B. C. Sousa, L. G. Dias, A. C. A. Veloso, L. Estevinho, A. M. Peres and A. A. S. C. Machado, Talanta, 2014, 128, 284–292 CrossRef CAS PubMed.
- S. Faal, M. Loghavi and S. Kamgar, Measurement, 2019, 148, 106936 CrossRef.
- C. I. G. Tuberoso, I. Jerković, G. Sarais, F. Congiu, Z. Marijanović and P. M. Kuś, Food Chem., 2014, 145, 284–291 CrossRef CAS PubMed.
- G. Bergamo, S. K. T. Seraglio, L. V. Gonzaga, R. Fett, R. D. de Mello Castanho Amboni, C. O. Dias and A. C. O. Costa, J. Food Sci. Technol., 2019, 56, 2771–2777 CrossRef CAS PubMed.
- S. Castiglioni, M. Stefano, M. Pisani and P. Carloni, Int. J. Food Sci. Technol., 2018, 53, 571–581 CrossRef CAS.
- A. Di Rosa, A. Marino, F. Leone, G. Corpina, R. Giunta and V. Chiofalo, Sensors, 2018, 18, 4065 CrossRef PubMed.
- M. Oroian and S. Ropciuc, Comput. Electron. Agric., 2019, 157, 371–379 CrossRef.
- J. M. Ramón-Sierra, J. C. Ruiz-Ruiz and E. de la Luz Ortiz-Vázquez, Food Chem., 2015, 183, 43–48 CrossRef PubMed.
- H. G. Gika, G. A. Theodoridis, R. S. Plumb and I. D. Wilson, J. Pharm. Biomed. Anal., 2014, 87, 12–25 CrossRef CAS PubMed.
- I. A. A. da Silva, T. M. S. da Silva, C. A. Camara, N. Queiroz, M. Magnani, J. S. de Novais, L. E. B. Soledade, E. de O. Lima, A. L. de Souza and A. G. de Souza, Food Chem., 2013, 141, 3552–3558 CrossRef PubMed.
- A. Michalkiewicz, M. Biesaga and K. Pyrzynska, J. Chromatogr. A, 2008, 1187, 18–24 CrossRef CAS PubMed.
- Y. Li, Y. Jin, S. Yang, W. Zhang, J. Zhang, W. Zhao, L. Chen, Y. Wen, Y. Zhang, K. Lu, Y. Zhang, J. Zhou and S. Yang, J. Chromatogr. A, 2017, 1499, 78–89 CrossRef CAS PubMed.
- S. K. T. Seraglio, A. C. Valese, H. Daguer, G. Bergamo, M. S. Azevedo, L. V. Gonzaga, R. Fett and A. C. O. Costa, Food Res. Int., 2016, 87, 60–67 CrossRef CAS PubMed.
- U. M. Gašić, M. M. Natić, D. M. Mišić, D. V. Lušić, D. M. Milojković-Opsenica, Ž. L. Tešić and D. Lušić, J. Food Compos. Anal., 2015, 44, 128–138 CrossRef.
- P. A. S. Tette, L. R. Guidi, E. M. A. F. Bastos, C. Fernandes and M. B. A. Gloria, Food Chem., 2017, 229, 527–533 CrossRef CAS PubMed.
- N. L. Hungerford, S. J. Carter, S. R. Anuj, B. L. L. Tan, D. Hnatko, C. L. Martin, E. Sharma, M. Yin, T. T. P. Nguyen and K. J. Melksham, Toxins, 2019, 11, 726 CrossRef CAS PubMed.
- N. Stanek, P. Kafarski and I. Jasicka-Misiak, J. Chromatogr. A, 2019, 1598, 209–215 CrossRef CAS PubMed.
- I. K. Karabagias, M. V Vavoura, A. Badeka, S. Kontakos and M. G. Kontominas, Food Anal. Methods, 2014, 7, 2113–2121 CrossRef.
- V. Vasić, U. Gašić, D. Stanković, D. Lušić, D. Vukić-Lušić, D. Milojković-Opsenica, Ž. Tešić and J. Trifković, Food Chem., 2019, 274, 629–641 CrossRef PubMed.
- Z. Jandrić, S. A. Haughey, R. D. Frew, K. McComb, P. Galvin-King, C. T. Elliott and A. Cannavan, Food Chem., 2015, 189, 52–59 CrossRef PubMed.
- U. Gašić, S. Kečkeš, D. Dabić, J. Trifković, D. Milojković-Opsenica, M. Natić and Z. Tešić, Food Chem., 2014, 145, 599–607 CrossRef PubMed.
- J. Zhou, L. Yao, Y. Li, L. Chen, L. Wu and J. Zhao, Food Chem., 2014, 145, 941–949 CrossRef CAS PubMed.
- S. Ouchemoukh, N. Amessis-Ouchemoukh, M. Gómez-Romero, F. Aboud, A. Giuseppe, A. Fernández-Gutiérrez and A. Segura-Carretero, LWT--Food Sci. Technol., 2017, 85, 460–469 CrossRef CAS.
- Z. Jandrić, R. D. Frew, L. N. Fernandez-Cedi and A. Cannavan, Food Control, 2017, 72, 189–197 CrossRef.
- M. Oroian and S. Ropciuc, Comput. Electron. Agric., 2017, 138, 148–156 CrossRef.
- G. M. Lo Dico, A. Ulrici, A. Pulvirenti, G. Cammilleri, A. Macaluso, A. Vella, V. Giaccone, G. Lo Cascio, S. Graci and M. Scuto, J. Food Compos. Anal., 2019, 82, 103225 CrossRef CAS.
- H. Kawashima, M. Suto and N. Suto, Food Chem., 2019, 289, 49–55 CrossRef CAS PubMed.
- M. Suto, H. Kawashima and N. Suto, J. Chromatogr. A, 2019, 1608, 460421 CrossRef CAS PubMed.
- I. K. Karabagias, A. Badeka, S. Kontakos, S. Karabournioti and M. G. Kontominas, Food Res. Int., 2014, 55, 363–372 CrossRef CAS.
- I. K. Karabagias, A. Badeka, S. Kontakos, S. Karabournioti and M. G. Kontominas, Food Chem., 2014, 146, 548–557 CrossRef CAS PubMed.
- I. K. Karabagias, A. V Badeka, S. Kontakos, S. Karabournioti and M. G. Kontominas, Food Chem., 2014, 165, 181–190 CrossRef CAS PubMed.
- P. Silva, J. Freitas, C. L. Silva, R. Perestrelo, F. M. Nunes and J. S. Câmara, Food Control, 2017, 73, 1176–1188 CrossRef CAS.
- N. M. Mădaş, L. A. Mărghitaş, D. S. Dezmirean, V. Bonta, O. Bobiş, M.-L. Fauconnier, F. Francis, E. Haubruge and K. B. Nguyen, Foods, 2019, 8, 445 CrossRef PubMed.
- I. K. Karabagias, A. Badeka and M. G. Kontominas, Microchem. J., 2020, 152, 104263 CrossRef CAS.
- A. C. V. da Costa, J. M. B. Sousa, T. K. A. Bezerra, F. L. H. da Silva, G. M. Pastore, M. A. A. P. da Silva and M. S. Madruga, LWT, 2018, 94, 198–207 CrossRef.
- A. Neggad, F. Benkaci-Ali, Z. Alsafra and G. Eppe, Chem. Biodiversity, 2019, 16, e1900267 CrossRef CAS PubMed.
- P. de L. M. da Silva, L. S. de Lima, Í. K. Caetano and Y. R. Torres, Food Res. Int., 2017, 102, 536–543 CrossRef PubMed.
- M. Juan-Borrás, E. Domenech, M. Hellebrandova and I. Escriche, Food Res. Int., 2014, 60, 86–94 CrossRef.
- I. Escriche, L. Sobrino-Gregorio, A. Conchado and M. Juan-Borrás, Food Chem., 2017, 226, 61–68 CrossRef CAS PubMed.
- F. Tanleque-Alberto, M. Juan-Borrás and I. Escriche, Food Chem., 2019, 277, 543–553 CrossRef CAS PubMed.
- N. Gerhardt, M. Birkenmeier, S. Schwolow, S. Rohn and P. Weller, Anal. Chem., 2018, 90, 1777–1785 CrossRef CAS.
- X. Wang, K. M. Rogers, Y. Li, S. Yang, L. Chen and J. Zhou, J. Agric. Food Chem., 2019, 67, 12144–12152 CrossRef CAS PubMed.
- A. Verzera, G. Tripodi, C. Condurso, G. Dima and A. Marra, Food Control, 2014, 39, 237–243 CrossRef CAS.
- M. Moniruzzaman, I. Rodríguez, M. Ramil, R. Cela, S. A. Sulaiman and S. H. Gan, Talanta, 2014, 129, 505–515 CrossRef CAS PubMed.
- S. Seisonen, E. Kivima and K. Vene, Food Chem., 2015, 169, 34–40 CrossRef CAS.
- M. S. Azevedo, S. K. T. Seraglio, G. Rocha, C. B. Balderas, M. Piovezan, L. V Gonzaga, D. de B. Falkenberg, R. Fett, M. A. L. de Oliveira and A. C. O. Costa, Food Control, 2017, 78, 383–392 CrossRef CAS.
- A. Pascual-Maté, S. M. Osés, G. L. Marcazzan, S. Gardini, M. A. Fernández Muiño and M. Teresa Sancho, J. Food Compos. Anal., 2018, 74, 34–43 CrossRef.
- M. S. Azevedo, S. K. T. Seraglio, G. Rocha, C. B. Balderas, M. Piovezan, L. V Gonzaga, D. de B. Falkenberg, R. Fett, M. A. L. de Oliveira and A. C. O. Costa, Food Control, 2017, 78, 383–392 CrossRef CAS.
- I. Escriche, L. Sobrino-Gregorio, A. Conchado and M. Juan-Borrás, Food Chem., 2017, 226, 61–68 CrossRef CAS.
- F. Tanleque-Alberto, M. Juan-Borrás and I. Escriche, Food Chem., 2019, 277, 543–553 CrossRef CAS PubMed.
- A. Klancnik, M. Kovac, N. Toplak, S. Piskernik and B. Jersek, Polymerase Chain React., Rijeka: Intech, cop, 2012, pp. 195–220, Search PubMedhttps://www.intechopen.com/books/polymerase-chain-reaction/pcr-in-food-analysis.
- I. Bruni, A. Galimberti, L. Caridi, D. Scaccabarozzi, F. De Mattia, M. Casiraghi and M. Labra, Food Chem., 2015, 170, 308–315 CrossRef CAS PubMed.
- Y. Wu, Y. Yang, M. Liu, B. Wang, M. Li and Y. Chen, J. AOAC Int., 2017, 100, 744–752 CrossRef CAS PubMed.
- S. Soares, L. Grazina, J. Costa, J. S. Amaral, M. B. P. P. Oliveira and I. Mafra, Food Control, 2018, 86, 367–373 CrossRef CAS.
- S. P. Kek, N. L. Chin, S. W. Tan, Y. A. Yusof and L. S. Chua, Food Control, 2017, 78, 150–159 CrossRef CAS.
- S. Soares, L. Grazina, I. Mafra, J. Costa, M. A. Pinto, H. P. Duc, M. B. P. P. Oliveira and J. S. Amaral, Food Res. Int., 2018, 105, 686–693 CrossRef CAS PubMed.
- S. Soares, J. S. Amaral, M. B. P. P. Oliveira and I. Mafra, Food Control, 2015, 48, 130–136 CrossRef CAS.
- S. P. Kek, N. L. Chin, S. W. Tan, Y. A. Yusof and L. S. Chua, Int. J. Food Sci. Technol., 2018, 53, 2490–2499 CrossRef CAS.
- V. J. Utzeri, A. Ribani and L. Fontanesi, Food Control, 2018, 91, 294–301 CrossRef CAS.
- E. I. Geana and C. T. Ciucure, Food Chem., 2020, 306, 125595 CrossRef CAS PubMed.
- E. Kivima, A. Seiman, R. Pall, E. Sarapuu, K. Martverk and K. Laos, Proc. Est. Acad. Sci., 2014, 63, 183–192 CrossRef CAS.
- L. Lenhardt, I. Zeković, T. Dramićanin, M. D. Dramićanin and R. Bro, Appl. Spectrosc., 2014, 68, 557–563 CrossRef CAS PubMed.
- Z. Gan, Y. Yang, J. Li, X. Wen, M. Zhu, Y. Jiang and Y. Ni, J. Food Eng., 2016, 178, 151–158 CrossRef CAS.
- M. Sahlan, S. Karwita, M. Gozan, H. Hermansyah, M. Yohda, Y. J. Yoo and D. K. Pratami, Vet. World, 2019, 12, 1304–1310 Search PubMed.
- A. P. Sobolev, S. Circi and L. Mannina, Advances in nuclear magnetic resonance spectroscopy for food authenticity testing, Elsevier Ltd, 2016 Search PubMed.
- M. Spiteri, E. Jamin, F. Thomas, A. Rebours, M. Lees, K. M. Rogers and D. N. Rutledge, Food Chem., 2015, 189, 60–66 CrossRef CAS.
- E. Schievano, C. Finotello, J. Uddin, S. Mammi and L. Piana, J. Agric. Food Chem., 2016, 64, 3645–3652 CrossRef CAS.
- E. Schievano, M. Stocchero, V. Zuccato, I. Conti and L. Piana, Food Chem., 2019, 288, 96–101 CrossRef CAS.
- E. Schievano, M. Sbrizza, V. Zuccato, L. Piana and M. Tessari, Food Chem., 2020, 309, 125788 CrossRef CAS.
- J. Bong, K. M. Loomes, R. C. Schlothauer and J. M. Stephens, Food Chem., 2016, 192, 1006–1014 CrossRef CAS.
- A. Guelpa, F. Marini, A. du Plessis, R. Slabbert and M. Manley, Food Control, 2017, 73, 1388–1396 CrossRef CAS.
- R. Consonni, F. Bernareggi and L. R. Cagliani, Food Control, 2019, 98, 133–140 CrossRef CAS.
- P. Pohl, A. Bielawska-Pohl, A. Dzimitrowicz, P. Jamroz, M. Welna, A. Lesniewicz and A. Szymczycha-Madeja, TrAC, Trends Anal. Chem., 2017, 93, 67–77 CrossRef CAS.
- M. Madejczyk and D. Baralkiewicz, Anal. Chim. Acta, 2008, 617, 11–17 CrossRef CAS PubMed.
- M. Jovetić, J. Trifković, D. Stanković, D. Manojlović and D. Milojković-Opsenica, J. AOAC Int., 2017, 100, 862–870 CrossRef.
- S. Squadrone, P. Brizio, C. Stella, S. Pederiva, F. Brusa, P. Mogliotti, A. Garrone and M. C. Abete, J. Trace Elem. Med. Biol., 2020, 126556 CrossRef CAS PubMed.
- S. Đ. Mračević, M. Krstić, A. Lolić and S. Ražić, Microchem. J., 2020, 152, 104420 CrossRef.
- P. Pohl, I. Sergiel and H. Stecka, Crit. Rev. Anal. Chem., 2009, 39, 276–288 CrossRef CAS.
- J. Atanassova, D. Pavlova, M. Lazarova and L. Yurukova, Biol. Trace Elem. Res., 2016, 173, 247–258 CrossRef CAS PubMed.
- P. Pohl, TrAC, Trends Anal. Chem., 2009, 28, 117–128 CrossRef CAS.
- J. Kováčik, J. Grúz, O. Biba and J. Hedbavny, Environ. Sci. Pollut. Res., 2016, 23, 4531–4540 CrossRef PubMed.
- S. Squadrone, P. Brizio, C. Stella, M. Mantia, S. Pederiva, F. Brusa, P. Mogliotti, A. Garrone and M. C. Abete, Environ. Sci. Pollut. Res., 2020, 1–12 Search PubMed.
- D. A. Magdas, F. Guyon, R. Puscas, A. Vigouroux, L. Gaillard, A. Dehelean, I. Feher and G. Cristea, Food Chem., 2020, 334, 127599 CrossRef PubMed.
- D. A. Magdas, O. Marincaş, G. Cristea, I. Feher and N. Vedeanu, Environ. Chem., 2020, 17, 148–157 CrossRef CAS.
- S. A. Drivelos, G. P. Danezis, S. A. Haroutounian and C. A. Georgiou, Food Chem., 2016, 213, 238–245 CrossRef CAS PubMed.
- D. A. Magdas, I. Feher, G. Cristea, C. Voica, A. Tabaran, M. Mihaiu, D. V. Cordea, V. A. Bâlteanu and S. D. Dan, Food Chem., 2019, 277, 307–313 CrossRef CAS PubMed.
- Z. Zhao, L. Chen, F. Liu, F. Zhou, J. Peng and M. Sun, Sensors, 2020, 20, 1878 CrossRef CAS.
- Y. Fiamegos, C. Dumitrascu, M. Ghidotti and M. B. De la Calle Guntinas, Anal. Bioanal. Chem., 2020, 412, 463–472 CrossRef CAS PubMed.
- Z. Sajtos, P. Herman, S. Harangi and E. Baranyai, Microchem. J., 2019, 149, 103968 CrossRef CAS.
- P. Zhang, C. A. Georgiou and V. Brusic, Briefings Bioinf., 2018, 19, 524–536 CAS.
- M. Oroian, A. Prisacaru, E. C. Hretcanu, S. G. Stroe, A. Leahu and A. Buculei, Int. J. Food Prop., 2015, 19, 1825–1836 CrossRef.
- H. Chen, C. Fan, Q. Chang, G. Pang, X. Hu, M. Lu and W. Wang, J. Agric. Food Chem., 2014, 62, 2443–2448 CrossRef CAS.
- I. K. Karabagias, A. P. Louppis, A. Badeka, C. Papastephanou and M. G. Kontominas, Eur. Food Res. Technol., 2019, 245, 1939–1949 CrossRef CAS.
- N. Czipa, L. Alexa, C. J. C. Phillips and B. Kovács, Eur. Food Res. Technol., 2018, 244, 1439–1445 CrossRef CAS.
- A. F. Shadan, N. A. Mahat, W. A. Wan Ibrahim, Z. Ariffin and D. Ismail, J. Forensic Sci., 2018, 63, 80–85 CrossRef PubMed.
- I. K. Karabagias, A. P. Louppis, S. Kontakos, C. Papastephanou and M. G. Kontominas, Eur. Food Res. Technol., 2017, 243, 101–113 CrossRef CAS.
- X. Zhou, M. P. Taylor, H. Salouros and S. Prasad, Sci. Rep., 2018, 8, 1–11 Search PubMed.
- G. P. Danezis, A. S. Tsagkaris, F. Camin, V. Brusic and C. A. Georgiou, TrAC, Trends Anal. Chem., 2016, 85, 123–132 CrossRef CAS.
- L. Bontempo, F. Camin, L. Ziller, M. Perini, G. Nicolini and R. Larcher, Measurement, 2017, 98, 283–289 CrossRef.
- V. Vasić, S. Đurđić, T. Tosti, A. Radoičić, D. Lušić, D. Milojković-Opsenica, Ž. Tešić and J. Trifković, Food Chem., 2020, 305, 125457 CrossRef PubMed.
- A. Schellenberg, S. Chmielus, C. Schlicht, F. Camin, M. Perini, L. Bontempo, K. Heinrich, S. D. Kelly, A. Rossmann and F. Thomas, Food Chem., 2010, 121, 770–777 CrossRef CAS.
- S. She, L. Chen, H. Song, G. Lin, Y. Li, J. Zhou and C. Liu, Food Chem., 2019, 272, 580–585 CrossRef CAS.
- O. R. Dinca, R. E. Ionete, R. Popescu, D. Costinel and G. L. Radu, Food Anal. Methods, 2015, 8, 401–412 CrossRef.
- I. K. Karabagias, G. Casiello, S. Kontakos, A. P. Louppis, F. Longobardi and M. G. Kontominas, Int. J. Food Sci. Technol., 2016, 51, 2460–2467 CrossRef CAS.
- V. Berriel and C. H. Perdomo, Heliyon, 2019, 5, e01228 CrossRef PubMed.
- E.-I. Geană, C. T. Ciucure, D. Costinel and R. E. Ionete, Food Control, 2020, 109, 106919 CrossRef.
| This journal is © The Royal Society of Chemistry 2021 |