Open Access Article
Open Access ArticlePotential repurposed SARS-CoV-2 (COVID-19) infection drugs
Gamal El-Din A. Abuo-Rahma *a,
Mamdouh F. A. Mohamed
*a,
Mamdouh F. A. Mohamed b,
Tarek S. Ibrahimcd,
Mai E. Shomana,
Ebtihal Samire and
Rehab M. Abd El-Bakyfg
b,
Tarek S. Ibrahimcd,
Mai E. Shomana,
Ebtihal Samire and
Rehab M. Abd El-Bakyfg
aDepartment of Medicinal Chemistry, Faculty of Pharmacy, Minia University, 61519 Minia, Egypt. E-mail: gamal.aborahama@mu.edu.eg; Tel: +20 1003069431
bDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Sohag University, 82524 Sohag, Egypt
cDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
dDepartment of Pharmaceutical Organic Chemistry, Faculty of Pharmacy, Zagazig University, Zagazig, 44519, Egypt
ePhysical Chemistry, Department of Analytical Chemistry, Faculty of Pharmacy, Deraya University, Minia 11566, Egypt
fDepartment of Microbiology and Immunology, Faculty of Pharmacy, Minia University, 61519 Minia, Egypt
gDepartment of Microbiology and Immunology, Faculty of Pharmacy, Deraya University, Minia 11566, Egypt
First published on 17th July 2020
Abstract
The global outbreak of COVID-19 viral infection is associated with the absence of specific drug(s) for fighting this viral infection. About 10 million people are already infected, about 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths all over the world to date. Great efforts have been made to find solutions for this viral infection, either vaccines, monoclonal antibodies, or small molecule drugs; this can stop the spread of infection to avoid the expected human, economic and social catastrophe associated with this infection. In the literature and during clinical trials in hospitals, several FDA approved drugs for different diseases have the potential to treat or reduce the severity of COVID-19. Repurposing of these drugs as potential agents to treat COVID-19 reduces the time and cost to find effective COVID-19 agents. This review article summarizes the present situation of transmission, pathogenesis and statistics of COVID-19 in the world. Moreover, it includes chemistry, mechanism of action at the molecular level of the possible drug molecules which are liable for redirection as potential COVID-19 therapeutic agents. This includes polymerase inhibitors, protease inhibitors, malaria drugs, lipid lowering statins, rheumatoid arthritis drugs and some miscellaneous agents. We offer research data and knowledge about the chemistry and biology of potential COVID-19 drugs for the research community in this field.
000 deaths all over the world to date. Great efforts have been made to find solutions for this viral infection, either vaccines, monoclonal antibodies, or small molecule drugs; this can stop the spread of infection to avoid the expected human, economic and social catastrophe associated with this infection. In the literature and during clinical trials in hospitals, several FDA approved drugs for different diseases have the potential to treat or reduce the severity of COVID-19. Repurposing of these drugs as potential agents to treat COVID-19 reduces the time and cost to find effective COVID-19 agents. This review article summarizes the present situation of transmission, pathogenesis and statistics of COVID-19 in the world. Moreover, it includes chemistry, mechanism of action at the molecular level of the possible drug molecules which are liable for redirection as potential COVID-19 therapeutic agents. This includes polymerase inhibitors, protease inhibitors, malaria drugs, lipid lowering statins, rheumatoid arthritis drugs and some miscellaneous agents. We offer research data and knowledge about the chemistry and biology of potential COVID-19 drugs for the research community in this field.
1 Introduction
This work aims to find a suitable vaccine or drug to save the global population promptly. One of the most promising strategies is drug repurposing, known as repositioning, reprofiling or redirecting strategies.1 This review article is focused on the chemistry and biology of drug molecules or drug groups that target diseases other than COVID-19 and may be useful to design possible drugs or drug combinations for treatment of COVID-19 viral infection.1.1 Background
Infectious diseases are those which are caused by microorganisms. Their importance comes from the degree and the extent of damage caused to the host cells and organs. Damage caused by microorganisms is due to their growth, multiplication or replication, their metabolic biproducts, distribution and their interference with functions of host systems.2 By the failure of immune system to eliminate the pathogenic organism, the infection can emerge.3 Emerging infectious disease usually appears in a certain time, in a given population of certain environment causing epidemics. Sometimes the infectious agent is highly contagious; spread rapidly resulting in the transfer of infection from one population to another and from one country to other causing pandemics. It was noticed in the last decades that many emerging infectious diseases which was controlled in the past, begins to reappear again. These reemerged infectious diseases found to be caused by the previously controlled pathogens after their acquisition of new properties due to gene mutation, genetic recombination, reassortment or the passage of the organism by many stages of adaptation to new environment and hosts.4 Many emerging infectious diseases were found to threaten the global public health causing pandemics such as SARS-COV (severe acute respiratory syndrome), MERS-COV (Middle East respiratory syndrome), hemorrhagic fever, Ebola, influenza and Zika virus infections. The impact of epidemics caused by the previous diseases depends on the number of the infected individuals in a certain time, mode of transmission, severity of cases and mortality rate.5On 12 December 2019, A Chinese patient (from Wuhan, Hubei province, china) suffering from severe respiratory disease was hospitalized followed by the admission of 1975 cases with the same symptoms until 25 January, 2020. By the increase in the number of cases with the same symptoms (fever, dry cough, fatigue and sore throat), samples were obtained from the bronchoalveolar lavage fluid from a patient (a worker at a seafood market) admitted to the central hospital of Wuhan, China for isolating the causative agent. Metagenomic RNA sequencing for the extracted RNA showed a new RNA virus belongs to coronavirdae that was named as 2019-nCOV on 7th January 2020. Then, it was renamed as SARS-COV-2 after testing its nucleotides' similarity to SARS-COV and MERS-COV that showed 89.1% of nucleotide identity between SARS-COV (was previously found in bats causing an outbreak in China, 2003) and the new virus.6
On 30 January 2020, the new coronavirus outbreak was declared as a public health emergency of international concern (PHEIC) which means that the new disease is an extraordinary event and is considered as a public health risk to other countries by the international spread of disease and require a coordinated international response.7 On 11 February 2020, World Health Organization (WHO) announced the official name for the new virus as coronavirus disease 2019 (COVID-19).8 This name was announced according to the guidelines of WHO set in 2015, which stated that naming new human disease should not refer to certain geographical area, animal, group of people to avoid the negative effects on travelling, tourism, trade, animal welfare, any cultural or national or professional or ethnic groups.9 On 11 March 2020, WHO officially changed the classification of COVID-19 from PHEIC to a pandemic disease which was the first pandemic since H1N1 pandemic in 2009.10
1.2 Epidemiology
As of 21 June 2020, WHO has reported that there are 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 894
894![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 711 confirmed cases of COVID-19 infection globally distributed in 6 different regions in 213 countries and territories. Out of the total number of the confirmed cases, active cases were 3
711 confirmed cases of COVID-19 infection globally distributed in 6 different regions in 213 countries and territories. Out of the total number of the confirmed cases, active cases were 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704
704![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 142 (364
142 (364![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 966 mild conditions (98.5%) and 54
966 mild conditions (98.5%) and 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 492 serious cases (1.4%)) and closed cases were 5
492 serious cases (1.4%)) and closed cases were 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 190
190![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 569 from which 4
569 from which 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724
724![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625 (91.1%) were recovered and 495
625 (91.1%) were recovered and 495![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 944 (9.5%) deaths,11 Fig. 1. Also, number of deaths among males were found to be more than that observed among females which may be attributed to females sex steroid concentrations and X chromosome diploidy that results in strong immune response observed by females,12 Fig. 2.
944 (9.5%) deaths,11 Fig. 1. Also, number of deaths among males were found to be more than that observed among females which may be attributed to females sex steroid concentrations and X chromosome diploidy that results in strong immune response observed by females,12 Fig. 2.
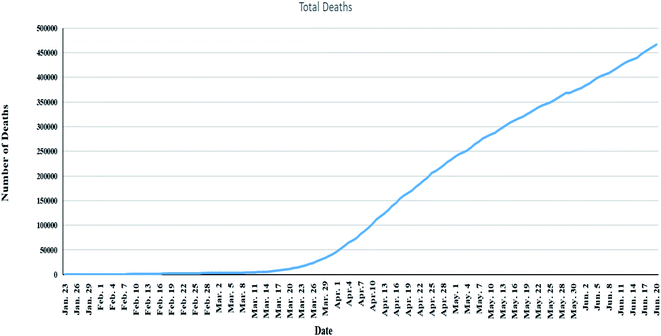 | ||
| Fig. 1 Global total deaths of COVID-19 starting from 23 January to 21 June 2020 (data from Worldometer).13 | ||
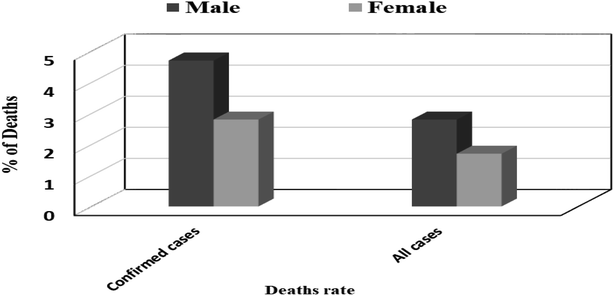 | ||
| Fig. 2 Global death rate among all cases and confirmed cases according to gender.13 | ||
1.3 Origin and transmission
COVID-19 or SARS-CoV-2 is a member of β-coronavirus, subgenus Sarbecovirus-Orthocoronavirinae subfamily.14,15 β-Coronaviruses were known to cause epidemics of severe acute respiratory illness such as epidemics caused by SARS-CoV in 2003 and MERS-CoV in 2012.16 Metagenomic RNA sequencing of the isolated strain, revealed 96.2% similarity to bat-CoV Ra TG13 and 89.1% similarity to SARS-CoV which suspected that it is a zoonotic disease and bat is the natural host of the virus. Also, previous findings suggested that COVID-19 uses angiotensin-converting enzyme-2 receptor (ACE2) similar to SARS-CoV. On the other side, the first isolated virus was isolated from a worker on a seafood market which did not contain bats that indicated the probability of the presence of some other intermediate hosts.17 Lui et al., reported that protein sequences alignment and genome sequencing showed that many species shared similar residues of receptors such as turtles, pangolins and snakes.18 Another study done by Wrapp et al.19 supported the probability of the presence of more than one intermediate host. As they revealed that binding affinity of SARS-CoV-2 S protein to ACE2 is much stronger than that observed by SARS-CoV. Human to human transmission was firstly reported on 30 January 2020 when a husband of a confirmed COVID-19 case tested positive for SARS-CoV-2 with no history of travelling outside USA.20 Many studies reported that 85% of human to human transfer occurs within social events and among family members. In addition, transmission of SARS-CoV-2 infection among health care team was found to be 3.8% of cases which is lower than that reported in SARS-COV in 2003 while majority of cases were infected from their families due to the long period of contact with them.6,10,21,221.4 COVID-19 (SARS-CoV-2) structure
SARS-CoV-2 viruses are enveloped positive sense RNA viruses with helical symmetrical nucleocapsid. Coronaviruses are characterized by their specific features of having club-shaped spike projections on their surfaces. Coronavirus particles have 4 main structural proteins which are: Spike (S) protein (trimetric S glycoprotein) is a class I fusion protein which is activated by human proteases and cleaved at S1/S2 containing receptor binding domain (RBD) and at S2′ portion responsible for virus fusion with cell membrane.23 M protein (∼25–30 kDa) gives the virus its shape and can adapt for 2 different conformations allowing it to promote membrane curvature to bind the nucleocapsid. E protein (∼8–12 kDa) is a transmembrane protein. It was found that viruses without E protein have no lethal action. It has a role in the assembly and the release of viruses. N protein is the protein forming nucleocapsid with high affinity for viral RNA. It has a role in the packaging of encapsidated genome to viral particles.24–26In addition, the virus has hemagglutination-esterase (HE) dimer in their structure which binds to sialic acid and shows esterase activity to facilitate viral S protein cell entry and viral spread.27
1.5 SARS-CoV-2 life cycle and pathogenesis
Viral attachment was initiated by binding S protein to ACE2 receptor. S protein is cleaved into S1, containing receptor binding site, which binds to peptidase domain of ACE2 receptor and S2 which is responsible for membrane fusion. After the binding of S protein with ACE2 receptor, they undergo conformational changes by pH-dependent cysteine protease cathepsin L. followed by fusion of viral envelope with the wall of endosome.28 Another way of entry depends on direct proteolytic cleavage of transmembrane protease 2 (TMPRSS2) to ACE2 receptor and the activation of S protein followed by the fusion of viral envelope with the host cell membrane and the passage of nucleocapsid into the cytoplasm and the release of viral genome.29Viral genomes act as mRNA. Translation was employed to the two third of the genome containing (open reading frame) ORF1a and ORF1b into polyproteins pp1a and pp1ab. Polyproteins with their proteases (PLpro and 3CLpro) were cleaved into 16 non-structural proteins forming replicase–transcriptase complex (RTC). The main protein of RTC is RNA-dependent RNA polymerase (RdRp) which mediates synthesis of negative sense subgenomic RNA from positive sense mRNA and the transcription of negative sense subgenomic RNA into positive sense mRNA and the replication of the positive mRNA to become the genome of the viral particles.30
The remaining part of the genome following ORF is translated into the structural proteins (S, E, M and N proteins) in the endoplasmic reticulum. Structural proteins move to Golgi intermediate compartment where M protein direct protein–protein interaction for protein assembly forming viral particles. Viral particles transferred by exocytosis using secretory vesicles for release.30
The peak viral load of patients with COVID-19 occurs during the first week of illness and gradually decreases over the second week which explained why SARS-CoV-2 is highly infectious and showed high transmissibility during the first week of getting infection. It was found that the severity of symptoms is correlated with age. Older adults showed more severe symptoms due to their lower immunity, presence of other diseases such as cardiovascular diseases, diabetes, pulmonary diseases, cancer or other diseases affecting their overall immunity31 and high expression of ACE2 receptors,32 Fig. 3.
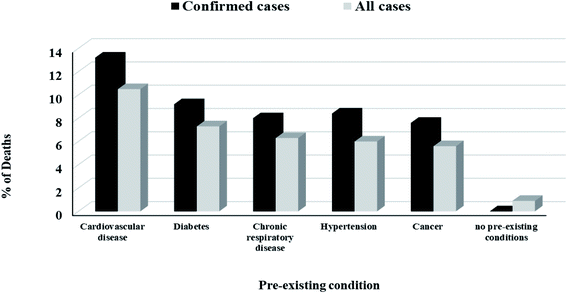 | ||
| Fig. 3 Death rate among all cases and confirmed cases according to the presence of pre-existing disease.13 | ||
1.6 Response to COVID-19 infection
Response to SARS-CoV-2 infection was found to have 3 stages depending on the overall immunity of patients. The first is asymptomatic stage with or without detectable virus. The second is symptomatic stage with mild to moderate symptoms with detectable virus. The third is symptomatic stage with severe complications, high viral load and marked damage to organs such as lung, intestine and kidney.33 Clinical manifestation of SARS-CoV-2 ranged from mild symptoms to life-threatening complications. Main symptoms are fever (88% of patients), dry cough (67% of patients), fatigue (38% of patients) and sore throat (14% of patients). Non-respiratory symptoms such as diarrhea are challenging to find. By time, patients develop shortness of breath (an average of eight days) followed by lymphopenia and clinical pneumonia supported with CT findings (characteristic pulmonary ground glass opacity changes on chest). Hypoxic respiratory failure may develop that needs ICU admission. Mechanical ventilation due to refractory hypoxia is required in 10% of patients.34According to previous data obtained during previous outbreaks caused by coronaviruses, innate immune response plays a critical role in the protection or the destructive response to the infection. SARS-CoV-2 was found to induce delayed type I IFN (interferon) and increase the influx of neutrophils and macrophages which are the sources of pro-inflammatory mediators (cause inflammation and further damage) with the loss of viral replication control. In addition, adaptive immune response was initiated by antigen presenting cell (APC) through their MHC class I and MHC class II by inducing CD8+ T cell response more frequently (to kill virus infected cells) than CD4+ T cell response which leads to the increase in the severity of disease and further damage to lung cells. In addition, humoral immunity is stimulated for the production of specific IgM at day 9 after disease onset and seroconversion to IgG by week 2.
Long incubation period of SARS-CoV-2 virus may be attributed to the time taken by the virus to evade immune detection mechanisms and to suppress immune response.17,35,36 Some patients remain viral positive or suffer relapse after their discharge from the hospital which indicates that some people may not develop immunity against SAR-CoV-2.37
Statistics results showed variation of number of deaths with age, Fig. 4. The rate of infection in the age range of 0 to 9 years old is very low. This may due to strong immune response or cross reactivity between vaccines of the routine childhood immunization schedule and other viral genus. Some studies showed that infants gain high degree of protection because of their extended immunization program. Liniger et al.38 showed that measles vaccine provides neutralizing antibodies against SARS-CoV and HIV. In addition, it was found that intralesional administration of MMR vaccine results in the improvement of cases suffering from warts caused by human papilloma virus. Also, Bordetella pertussis (a vaccine against whooping cough disease) and BCG (a vaccine against tuberculosis diseases) vaccines were reported to give rise to neutralizing antibodies against some respiratory viruses by the production of local cytokines in lungs protecting lungs from damage. This situation may be explained by the ability of vaccines to stimulate CD4+ T helper1 cells resulting in the secretion of different types of cytokines including γ-interferon, IL-2, IL-12 activating cytotoxic NK cells and stimulating CD8+ T cells for killing virus infected cells.39–41 Another explanation for child response to infection may be attributed to the trained immunity or innate immunity memory in which innate immune system stores information about the pathogen causing primary infection or after vaccination to respond rapidly and strongly after the subsequent exposure to the same or some unrelated pathogens. Many studies reported metabolic, epigenetic and transcriptional reprogramming for specific innate immune cells such as monocytes, macrophages and NK cells regarding strong non-specific vaccination response.42 Also, it was reported that this memory is transferable especially after observing trained monocytes (monocytes have short-life span in circulation of one day) in the circulation of an individual after 3 months of BCG vaccination. Innate memory was explained by the occurrence of reprogramming at the level of progenitor cells of the innate immune cells and the ability of innate immune memory to transfer via hematopoietic stem and progenitor cells. On the other hand, the signals mediating the transfer of innate immune memory from the periphery to the bone marrow is not yet known in details and need further investigation.42–44
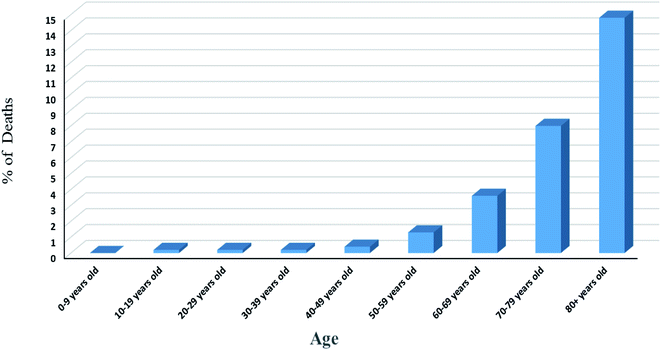 | ||
Fig. 4 Death rate among different age range according to data obtained from the first 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 672 confirmed cases in China.50 672 confirmed cases in China.50 | ||
Many studies reported the protective role of breastfeeding even after breastfeeding has been stopped. As it gives protection against infection by wide range of pathogens including respiratory pathogens in the long term which means that its protective role increase by the increase in the time of breastfeeding showing persistent protective effect.45,46 Agarwal et al.47 reported that macrophage cells represent 80% of the early milk and by the transfer of macrophages to the infant, they can be differentiated into dendritic cells that can stimulate T-lymphocytes giving protection against wide range of pathogens. Also, human milk contains pro-inflammatory cytokines in levels that can induce inflammation and defend against pathogens, fatty acids which have destructive effect on microbial membranes and viral envelope,48 lactoferrin, iron binding glycoprotein (belongs to transferrin family) and milk fat globule containing mucins which are known with their activity against bacteria, viruses and fungi.49
1.7 Potential interventions for treating or preventing COVID-19
There are no approved treatment or effective vaccines against SARS-CoV-2 till now but there is a great need for rapid development of antiviral agents and vaccines to stop the wide spread of infection and to save lives of the infected cases. There are many trials applied and showed useful results such as:Kim et al. produced both rSARS-CoV-S1 and rSARSCoV-S1fRS09 immunogens after the publication of the SARS-CoV-2-spike glycoprotein sequence and incorporate them in dissolvable microneedle array (MNA) for pre-clinical testing publication. MNA delivery of either rSARS-CoV-2-S1 or rSARS-CoV-2-S1fRS09 induced significant increases in antigen-specific antibodies responses as soon as week 2. So, it is considered as a promising immunization strategy against coronavirus infection.58
2 Repofiling of known antiviral agents in COVID-viruses
2.1 Polymerase inhibitors
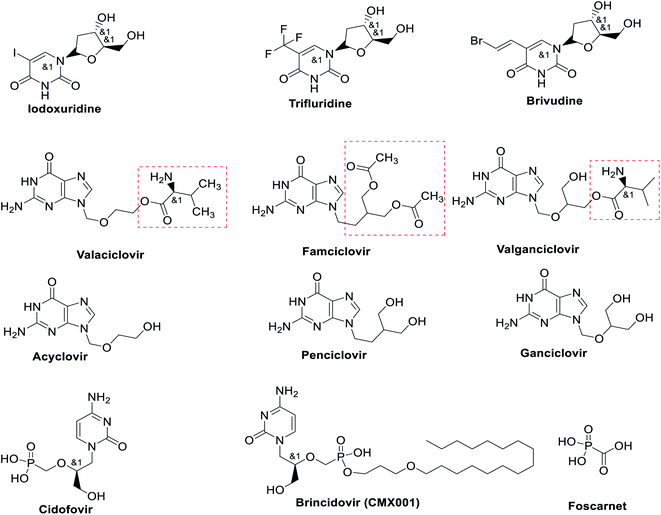
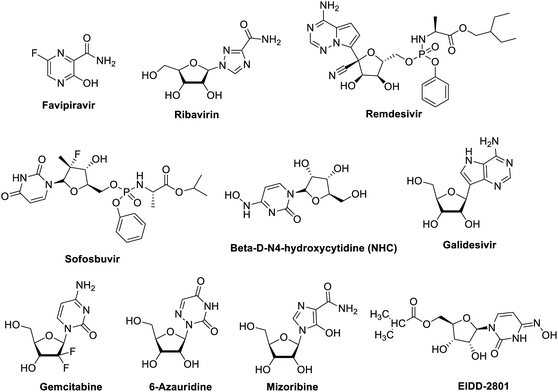
Polymerase inhibitors, in general, can be classified into two categories: nucleotide/nucleoside analogs and non-nucleoside inhibitors (pyrophosphate derivatives). Nucleotide and nucleoside analogue inhibitors are chemically synthesized analogues of purines and pyrimidines in which the heterocyclic ring or sugar moiety has been altered and include 5-substituted 2′-deoxyuridine analogs (idoxuridine, trifluridine, and brivudine) Fig. 5 and Table 1. They include arabinosyl nucleoside analogs (vidarabine) and acyclic guanosine analogs (acyclovir, ganciclovir, penciclovir, famciclovir, valaciclovir, and valganciclovir) (Fig. 5). Related to these drugs, cidofovir is a broad-spectrum nucleotide analog (acyclic nucleoside phosphonate), approved for treatment of HCMV retinitis in AIDS patients and used to treat many other DNA virus infections (e.g., HSV, adeno-, pox-, polyoma-, and papillomavirus infections) (Fig. 5).60,61| Antiviral agents | Infectious diseases | Reported mechanism of action | Status |
|---|---|---|---|
| Favipiravir | 2019-nCoV; influenza | Inhibits RdRp | • Approved for influenza in Japan |
| • Randomized trial for 2019-nCoV | |||
| (ChiCTR2000029544, ChiCTR2000029600) | |||
| Ribavirin | 2019-nCoV, MERS-CoV, SARS-CoV, RSV, HCV | Inhibits viral RNA synthesis and mRNA capping | • Approved for HCV and RSV |
| • Randomized trial for 2019-nCoV in combination a pegylated interferon (ChiCTR2000029387) | |||
| • Randomized trial for SARS (NCT00578825) | |||
| Penciclovir | 2019-nCoV | Inhibits RdRp | Approved for HSV |
| Remdesivir (GS-5734) | 2019-nCoV, MERS-CoV, SARS-CoV | Terminates the non-obligate chain | • Phase III for 2019-nCoV (NCT04252664, NCT04257656) |
| • Phase I for Ebola (NCT03719586) | |||
| Galidesivir (BCX4430) | Broad-spectrum (e.g. SARS-CoV, MERS-CoV, IAV) | Inhibits viral RNA polymerase function by terminating non-obligate RNA chain | • Phase I for yellow fever (NCT03891420) |
| • Phase I for Marburg virus (NCT03800173) | |||
| 6′-Fluorinated-aristeromycin analogues | Broad-spectrum (e.g. CoV, ZIKV, CHIKV) | Inhibits the activity of RdRp and host cell S-adenosyl-L-homocysteine hydrolase | Preclinical |
| Acyclovir fleximer analogues | HCoV-NL63, MERS-CoV | Doubly flexible nucleoside analogues inhibit RdRp | Preclinical |
Nucleoside analogs are prodrugs that mimic natural nucleosides and are used, in their triphosphate forms, as substrates for the synthesis of viral nucleic acid. This means that nucleoside analogs must not be only substrates for viral polymerases, they also need to be recognized and phosphorylated by host nucleo(s/t)ide kinases.60,61 Nucleotide and nucleoside analogue inhibitors (NIs) currently used to treat both acute and chronic viral infections. NIs exhibit a relatively high barrier to resistance emergence because the structural conservation of the polymerase targets binding site is high among virus families, and resistance mutations generally incur a fitness cost for the enzyme and the virus.62
For CoVs, amino acid conservation of the viral RdRp ranges from 70 to near 100% and is maintained across genera, suggesting NIs could potentially serve as broad-spectrum inhibitors of CoV infection.63 However, proofreading activity of nsp14-ExoN activity protects CoVs from many NIs effective against other RNA viruses.64,65 For effective inhibition of CoVs, an NI needs to either evade recognition by ExoN or undergo uptake into the elongating strand at a rate exceeding ExoN excision kinetics.
2.1.1.1 Favipiravir. Favipiravir, also known as T-705, pyrazinecarboxamide derivative, a guanine nucleoside analogue (Fig. 6 and Table 1) was originally developed by Toyama Chemical of Japan.66 In March 2015, the US Food and Drug Administration completed a phase III clinical trial studying the safety and efficacy of favipiravir in the treatment of influenza.67 Moreover, on March 15, 2020 the drug was approved in China for the treatment of influenza.68 Recently, favipiravir approved for use in clinical trials for treating coronavirus disease 2019 pneumonia.68 Its mechanism of actions is thought to be related to the selective inhibition of viral RNA-dependent RNA polymerase.69 Other research study suggests that favipiravir induces lethal RNA transversion mutations, producing a nonviable viral phenotype.70 Favipiravir is a prodrug that is metabolized to its ribofuranosyl-5′-triphosphate (favipiravir-RTP) form which act as purine mimetic binding to ATP- and GTP-binding sites on polymerase in a competitive manner.71–73 Notably, favipiravir is selective to viral cells, it does not inhibit RNA or DNA synthesis in mammalian cells therefor it is not toxic to them.74 Favipiravir can effectively inhibit the RNA-dependent RNA polymerase of RNA viruses such as influenza, Ebola, yellow fever, chikungunya, norovirus and enterovirus.75
In February 2020, favipiravir has been approved for a clinical trial as a drug to treat COVID-19.76,77 On March 17, Chinese officials suggested the drug had been effective in treating COVID-19 in Wuhan and Shenzhen. Recently, a study reported its activity against 2019-nCoV (EC50 = 61.88 μM in Vero E6 cells).78 Patients with 2019-nCoV are being recruited in randomized trials to evaluate the efficacy of favipiravir plus interferon-α (ChiCTR2000029600) and favipiravir plus baloxavir marboxil (an approved influenza inhibitor targeting the cap-dependent endonuclease) (ChiCTR2000029544). Moreover, A study on 80 patients comparing it to lopinavir/ritonavir found that it significantly reduced viral clearance time to 4 days, compared to 11 for the control group, and that 91.43% of patients had improved CT scans with few side effects.79,80 As of 23 March 2020, it seems that Japan and China have issued an export ban on the substance. Japan and China are the only countries in which favipiravir is produced and approved as a medical compound. Some Chinese pharmaceutical companies assure that export rights are still granted via international diplomatic means by the Chinese Ministry of Industry and Information Technology.
2.1.1.2 Ribavirin. Ribavirin (Fig. 6) is a synthetic nucleoside guanosine analogue and has broad-spectrum antiviral activity against multiple RNA viruses. It is approved for treating respiratory syncytial virus, hepatitis C and E virus, Lassa virus, and hantavirus infections. Although the coronavirus is an RNA virus, in theory favipiravir and ribavirin have some anti-coronavirus activity. However, coronaviruses express exonuclease (nsp14-ExoN) in nonstructural protein 14, and are common throughout the coronavirus family. The current research results show that nsp14-ExoN has an RNA proofing function.81 So, it is speculated that the coronavirus is resistant to nucleoside analogs. The results of in vitro experiments have shown that ribavirin has little antiviral effect on coronavirus. NIs effective against other RNA viruses.64 Synergistic activity against MERS-CoV of ribavirin combined with IFNα2b was observed in vitro and in rhesus macaques, suggesting that IFN increases the potency of ribavirin at lower, more tolerable concentrations.82,83 However, five critically ill MERS-CoV-positive patients who were treated with a combination of ribavirin and IFNα2b showed no clinical improvement.84 Treatment of 20 MERS patients with a combination of ribavirin and IFNα2a showed significantly improved survival at 14 days but not at 28 days,85 whereas treatment of MERS patients with a combination of IFNα2a or IFNβ1a and ribavirin yielded no survival benefit in another study.86 Thus, although ribavirin shows some efficacy in vitro, it does not provide clinical benefit to humans with SARS-CoV or MERS-CoV infections. Ribavirin approved for treating HCV and respiratory syncytial virus (RSV) that has been evaluated in patients with SARS and MERS, but its side effects such as anaemia may be severe at high doses87 and whether it offers sufficient potency against 2019-nCoV is uncertain. However, a fixed dose of the anti-HIV combination, lopinavir–ritonavir, is currently in clinical trials with umifenovir or ribavirin.88
2.1.1.3 Remdesivir (GS-5734). Remdesivir (Fig. 6), nucleoside analogue, is a phosphoramidate prodrug of the adenosine NI GS-441524 which have multiple mechanisms of action, including lethal mutagenesis, obligate or non-obligate chain termination, and perturbation of natural nucleotide triphosphate pools via inhibition of nucleotide biosynthesis.89 Remdesivir has demonstrated antiviral activity in vitro against several viral families of emerging infectious diseases including pneumoviridae, filoviridae, pneumoviridae, paramyxoviridae, and coronaviridae.63,90,91 Remdesivir has broad-spectrum activities against a diverse panel of RNA viruses such as Ebola virus (EBOV), Marburg, MERS-CoV, SARS-CoV, respiratory syncytial virus (RSV), Nipah virus (NiV), and Hendra virus under clinical development. The mechanism of remdesivir's anti-MERS-CoV activity is likely through premature termination of viral RNA transcription.92,93 A recent study reported that remdesivir inhibited 2019-nCoV (EC50 = 0.77 μM in Vero E6 cells),78 and a US patient with 2019-nCoV recovered after receiving intravenous remdesivir in January.94 The drug is currently being studied in phase III clinical trials in both China and USA. Two phase III trials were initiated in early February to evaluate intravenous remdesivir (200 mg on day 1 and 100 mg once daily for 9 days) in patients with 2019-nCoV (NCT04252664 and NCT04257656), with estimated completion dates in April 2020.95 Studies probing the interactions between remdesivir and the CoV replication machinery will likely yield crucial insights into how this NI circumvents or overcomes CoV proofreading activity, which can in turn be applied to modeling the development of new NIs and enhancing potency of existing NIs. Currently, remdesivir clinical trials are enrolling and are supported by the National Institutes of Health (NIH),96 USA and AIFA, Italy.97 The use of remdesivir in COVID-19 patients in a single-arm trial gave positive preliminary outcomes,98 and this in contrast to the result published by another group.99 However, the debate over remdesivir efficacy is still open, according to the reported preliminary results in the ACTT NIH clinical trial,96 the FDA has given remdesivir an emergency use authorization restricted to patients affected by sever COVID-19.97
2.1.1.4 Sofosbuvir. Sofosbuvir (Fig. 6), a nucleotide analogue hepatitis C virus NS5B polymerase inhibitor, is used to treat chronic hepatitis C as a component of a combination antiviral regimen. In addition, the European Medicines Agency's Committee for Medicinal Products for Human Use has recommended the approval of sofosbuvir for the treatment of chronic hepatitis C.100,101 The most important druggable targets of sofosbuvir is the RNA-dependent RNA polymerase (RdRp), where it is incorporated into RNA, and due to modifications at the 2′ position, inhibits further RNA chain extension and halts RNA replication, and sofosbuvir can inhibit RdRp of the hepatitis C virus, it acts as an RNA polymerase inhibitor by competing with natural ribonucleotides. As the hepatitis C virus and the coronavirus use a similar viral genome replication mechanism, sofosbuvir may also inhibit coronaviruses, including 2019-nCoV.102
2.1.1.5 Beta-D-N4-hydroxycytidine (NHC). NHC (Fig. 6) is a cytidine analogue with demonstrated potent, broad-spectrum antiviral activity against many viruses such as Venezuelan equine encephalitis virus (VEEV), influenza A virus (IAV), respiratory syncytial virus (RSV), chikungunya virus (CHIKV), influenza B virus (IBV), and CoVs. NHC exerts its antiviral effect primarily through mutagenesis of viral RNA serial passaging in the presence of NHC led to low level resistance for VEEV but not RSV, IAV, and bovine viral diarrhea virus, thus indicating a high resistance barrier.103–107 Potent anti-CoV activity of NHC was demonstrated for SARS-CoV and HCoV-NL63.106,108 Although the mechanism of CoV inhibition has not been determined, micromolar-range EC50s suggests that NHC like remdesivir may also have a novel way of interacting with the CoV replication. Its potent, broad-spectrum antiviral activity warrants further investigation for the treatment of CoV infections, either alone or in combination with other DAAs and immunomodulators. Moreover, the isopropyl ester prodrug of the ribonucleotide analogue, N4-hydroxycytidine (Fig. 6) with improved bioavailability was found to inhibit replication of human and bat SARS-Cov-2. This occurs in the airway epithelial cells in both mice and human. Clinical trials for the effectiveness of this drug are in progress.109
2.1.1.6 Other drugs. Galidesivir (BCX4430), Fig. 6, is an adenosine analogue that was originally developed for HCV, and has broad spectrum activity positive and negative sense RNA viruses. Galidesivir is currently in early-stage clinical studies evaluating its safety in healthy subjects and its efficacy against yellow fever, and has shown antiviral activities in preclinical studies against many RNA viruses, including SARS and MERS.87,110
Gemcitabine hydrochloride, Fig. 6, another NIs deoxycytidine analogue, is a chemotherapy drug that inhibits SARS-CoV and MERS-CoV, in addition to the uridine analogue 6-azauridine (Fig. 6) with activity against HCoV-NL63;106 and finally, mizoribine (Fig. 6), the immunosuppressant imidazole nucleoside, which inhibits SARS-CoV.87
2.2 Proteases inhibitors
The use of protease inhibitors for preventing coronavirus replication started with repurposing anti-HIV protease inhibitors. While the main residue in HIV protease is an aspartate residue,119 coronaviruses protease uses a cysteine residue for nucleophilic attack.120,121 The approach of using HIV protease inhibitors for older type of coronavirus SARS-CoV protease started with rational screening for old medicines including antivirals to block the proteolytic activity of SARS-CoV.120 Since the Mpro sequence of both types of coronavirus is conserved by 96% and homology studies showed they also share a very similar 3D structure, the use of drugs that block Mpro of SARS-CoV as potential SARS-CoV-2 protease inhibitors is rational.112
Actually, previously approved anti-HIV protease inhibitors; lopinavir and ritonavir (Fig. 7); were found to be promising candidates for tackling the older type of coronavirus, SARS-CoV, active site.122–124 Theoretically both lopinavir and ritonavir was bound efficiently to the same pocket in active sites of Mpro of both SARS-CoV and SARS-CoV-2.115 Unfortunately, a randomized clinical study performed on 199 hospitalized patients in China and USA did not prove that use of the anti-HIV drugs of clinical significance over the standard care introduced for SARS-CoV-2 affected patients.124,125 A similar contradiction between cell culture testing and in vivo screening results was reported earlier also with using alisporivir; is a cyclophilin inhibitor; against emerged coronaviruses SARS and MERS. Alisporivir inhibited both viruses in EC50 of about 3.6 mM but it failed to show any improvements of SARS infection in mouse model.126 Another anti-HIV protease inhibitor, nelfinavir, was repurposed for SARS infection. Nelfinavir showed a strong inhibitory activity on SARS-CoV replication127 and though it gave strong reduction in viral pathogenicity and viral antigen expression for SARS-CoV but no reports tested on SARS-CoV-2 yet.
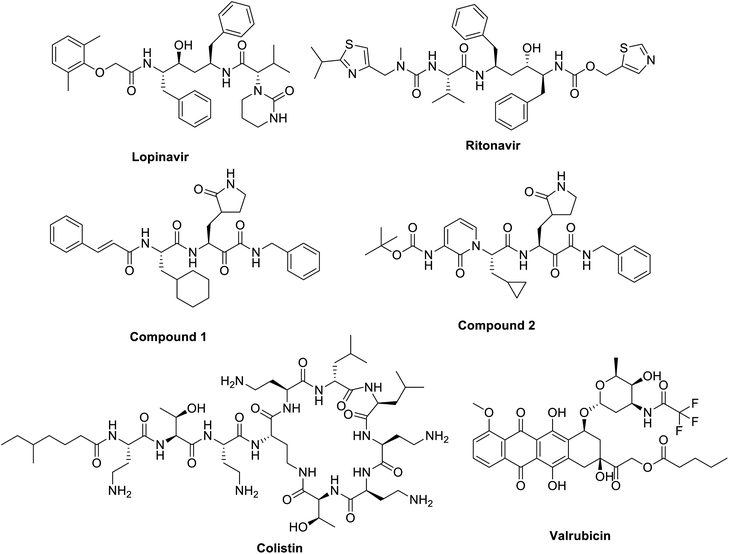 | ||
| Fig. 7 Structure of lopinavir and ritonavir, a-ketoamide protease 1 and 2, anti-HIV protease inhibitors. | ||
Based on the separated crystal structure of SARS-CoV-2, a German Laboratory112 was able to design a-ketoamide inhibitor (compound 2, Fig. 7) that successively inhibited SARS-CoV-2 viral replication in infected human Calu3 cells with an EC50 of 4–5 μM. The design is based on a previously designed SARS-CoV protease inhibitor introduced earlier this year (compound 1, Fig. 2). The ability of these a-ketoamides to form a bond on the Cys145 residue in the active site of SARS-CoV series forming a thiohemiketal offered an advantage over HIV protease inhibitors used lacking thiol reactivity.128 An additional advantage is the ability of these compounds to form two hydrogen bonds at the catalytic active sit of SARS-CoV-2 with Gly143, Cys129,145 but still the lack of animal testing raising a big question on the availability of use of such inhibitors for controlling COVID-19.
Additionally, a theoretical study using the crystal structure of the Mpro for SARS-CoV2, scientists tried to repurpose commercially available medicines to fit into the enzyme pocket. Surprisingly, antibiotics and chemotherapeutic agents named colistin and valrubicin (Fig. 7) showed tight binding with 9 and 7 hydrogen bonds formed with essential amino acid residues in Mpro active site including THR24, THR25, and THR26.115 Other theoretical studies investigating millions of small molecules as possible candidates for Mpro inhibitions are coming out lipoproteins112,130 but the lack of biological testing hinders the use of these molecules as potential Mpro inhibitors and a need for more evidence to support to support their use is still required.
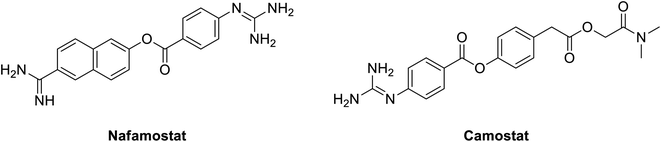
In 2016, nafamostat; Fig. 8, a blood thinner that works via serine protease inhibition approved in Japan for treatment for acute Pancreatitis, Fig. 4; was repurposed to inhibit middle east coronavirus MERS-CoV.135 Currently, it is also tested to prevent SARS-CoV-2 entry into human cells. Nafamostat can prevent the fusion of the envelope of the virus with host cell surface membranes, the first step in infection with the causative virus SARS-CoV-2. Similarly, another inhibitor of TMPRSS2, camostat, blocks SARS-CoV-2 infection of human lung cells, Fig. 8; it actually minimized entry of MERS-S, SARS-S and SARS-2-S proteins into lung cell lines.131 Nafamostat showed 10 times lower concentration in inhibiting viral membrane fusion compared to camostat.136 Generally, they both represent an interesting class of compounds that might be useful in our current fight against the COVID-19 endemic.
2.3 Malaria drugs in COVID-19
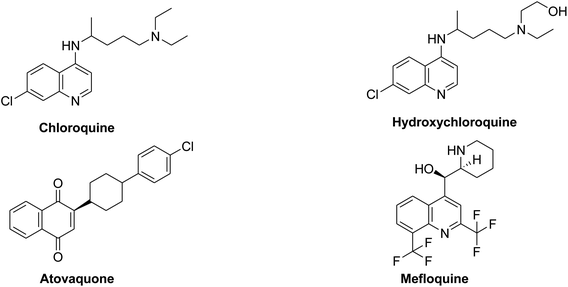
Chloroquine, hydroxychloroquine and the structurally related atovaquone and mefloquine, Fig. 9, are old known therapeutically used antimalarial drugs. Hydroxychloroquine, the hydroxyl analogue of chloroquine being synthesized in the mid of 20th century was introduced as a less toxic derivative than the parent chloroquine in animal studies. Moreover, these two drugs are known to have immunomodulatory effects; hence they have been used in rheumatoid arthritis or in lupus erythematous diseases.137A collective review about the possible in vivo and in vitro antiviral activity of malaria drugs concluded that utilization of antimalarial drugs against viral infection may be effective especially in cases of viral resistance and emergences.138 An in vitro study showed that The IC50 of chloroquine against SARS-CoV was 8.8 ± 1.2 μM which is less than the cytostatic activity; CC50 (261.3 ± 14.5 μM) with selectivity index 30. The IC50 of chloroquine for inhibition of SARS-CoV in vitro approximates the plasma concentrations of chloroquine reached during treatment of acute malaria. Addition of chloroquine to infected cultures could be delayed for up to 5 h post infection, without a significant drop-in antiviral activity. Authors concluded that chloroquine, an old antimalarial drug, may be considered for prompt use in the prevention and treatment of SARS-CoV infections.139 Also, another in vitro study resulted in important results where chloroquine can inhibit the replication and spread of CoV, and prevents CoV infection in newborn mice; this is considered a promising potential therapy for this resistant virus.140 Recent reports about chloroquine and hydroxychloroquine recommend them as SARS-CoV potential agents. In vitro research studies results showed that hydroxychloroquine is three times more potent than chloroquine in its effect on SARS-CoV-2 infected cells (EC50 = 0.72 and 5.47 μM). This proposal depends upon the immunomodulatory effect of chloroquine and hydroxychloroquine which may be useful in controlling the cytokines stress that occurs in patients with SARS-CoV-2.141 Vincent et al., (Vincent et al.)142 concluded that chloroquine is effective in preventing the spread of SARS-CoV in cell culture. This effect was found to occur in cells either treated with chloroquine prior or after SARS-CoV infection. This study indicated that chloroquine could elevate endosomal pH or can interfere with terminal glycosylation of the cellular receptor, angiotensin converting enzyme-2. These actions can affect virus receptor binding and hence repeal viral infection. A most recent study revealed that hydroxylchloroquine may contribute efficiently in inhibiting SARS-CoV-2 in vitro. It may attenuate inflammatory response associated with COVID-19 and hence it can compete with the infection if the toxicity profile has been established by clinical studies.18
It is noteworthy worth that the N-cinnamoyl analogues of chloroquine 3a, b, Fig. 10, are effective agents in vitro in relieving pneumocystis pneumonia associated with some pulmonary viral, bacterial, mycobacterial or parasitic infections. Compounds 3a and 3b will be further assessed in in vivo assays as potential potent anti-pneumocystis pneumonia.143
The mechanism of broad-spectrum antiviral of chloroquine and hydroxychloroquine is associated with suppressive effect on the production of the inflammatory mediators TNF-α and IL6. Authors in this study suggest hydroxychloroquine and analogues to have useful clinical applications in treatment of viral infections associated with inflammation or immune-hyperactivation. Moreover, a series of chloroquine analogues 4, Fig. 10 (ref. 144) can inhibit tumor susceptibility gene (TSG101) and also seems to inhibit viral replication by blocking late-stage viral activity, possibly after completion of viral protein synthesis. Inhibition of this gene could retard the movement of the virus to the cell surface and subsequently affect budding.145
Moreover, the antimalarial antiparasitic agent atovaquone (Fig. 9) acts via retardation of pyrimidine biosynthesis and consequently inhibits viral replication. In vitro study using human placental model showed that atovaquone can limit Zika virus infection and suggesting that it can serve as broad spectrum antiviral effect.146 Theoretically, computer-based design and screening proposed that chloroquine may act as SARS-CoV-2 entry blocker with S-score of 7.2639 kcal mol−1.147 Atovaquone showed a potential binding affinity with S-score of −0.8.449 kcal mol−1 This study states that atovaquone has the same scaffold features like chloroquine to elevate endosomal pH and interfere with ACE-2 glycosylation.147 In very recent research study, a model was used for 2019-nCoV research, results showed that the antimalarial mefloquine HCl, and the antiparasitic selamectin are potential agents for treating COVID-19 infection.148
Due to the geographical overlaps between malaria and viral-related diseases, antimalarial drugs represent additional modes and mechanisms of action as antiviral agents. The lack of new effective antiviral drugs has strengthened interest in the potential antiviral activity of antimalarial drugs.149 It is obvious that mechanism of action of chloroquine is not certain, it may act by elevation of endosomal pH or interference with terminal glycosylation of the cellular receptor, angiotensin converting enzyme-2.150 Chloroquine inhibits quinone reductase, a structural neighbor of UDP-N-acetylglucosamine-2-epimerase that is involved in the biosynthesis of sialic acid. Viruses can use sialic acid moiety as receptor.151,152
Chloroquine and hydroxylchloroquine are considered promising in treatment of COVID-19. Their previous effects on MERS-COV, the analogue of COVID-19 in addition to the recent studies about their effects153 recommend them as potential urgent solution to treat this viral infection on some inflammatory mediators involved in pneumonia associated with COVID-19 infections recommend them as potential urgent solution to treat this viral infection after completing the clinical studies. Also, medicinal chemists are invited to study the molecular target mechanism of chloroquine and its analogues as promising therapy for COVID-19 that enables development of newer better targeted derivatives with less side effects.
Many clinical trials and studies, in addition to in vitro and in vivo, are currently ongoing to validate the efficacy of chloroquine and hydroxychloroquine as effective treatment for SARS-COV-2. They are testing chloroquine if able to shorten SARS-CoV-2 disease course, mitigate inflammatory responses to infection, inhibit the exacerbation of pneumonia, improve lung imaging findings, and promote a virus negative conversion.154–158 On the 13th March 2020, the FDA approved chloroquine as a treatment for COVI-19 infection with specific precautions. While early studies suggested beneficial effects of chloroquine and hydroxylchloroquine, recent studies could not confirm this, but instead highlighted potential meaningful adverse events of chloroquine and hydroxylchloroquine in the treatment of COVID-19; this led to a temporary pause from May 23 to June 3 of the chloroquine and hydroxylchloroquine arm within the large, international Solidarity trial.159 After a reassessment of the evolving data the FDA revoked the emergency use authorization for chloroquine and hydroxylchloroquine on June 15.160 Therefore, we must pay the attention chloroquine and hydroxylchloroquine need to be administered with caution when treating COVID-19 infection to prevent the possible cardiovascular problems.160
2.4 The lipid lowering statins in COVID-viruses
Rosuvastatin (RSV), Fig. 11, is a statin FDA-approved to be used as lipid lowering agent, it acts through inhibition of HMG-CoA reductase enzyme consequently it lowers or cholesterol level. This molecule has the advantages of being inexpensive, safe and easily obtained.161 A recent study showed that rosuvastatin improves lung pathological changes by decreasing T helper cells Th2 and Th17-mediated cytokines where this action is not related to its lipid-decreasing activity.162Another study by Farag et al., in a structure-based drug design approach aiming at targeting COVID-19 virus revealed that rosuvastatin on docking along with COVID-19 virus Mpro substrate-binding pocket (PDB ID: 6LU7). Rosuvastatin showed outstanding binding affinity regarding free energy with S score of −12.3096 kcal mol−1. Concerning binding mode, it experienced hydrophobic interactions and hydrogen bonding with Gly143 and Glu166 amino acids.147 Although the study was totally computer-based, and need more validation studies before any clinical application, the study gives a starting point to consider the re-evaluation of statin against COVID-19.
Indeed, some research studies indicate that statins therapy is associated with a reduction in cardiovascular problems and mortality rates in patients of influenza and/or pneumonia.147 Totura et al.163 proposed that toll-like receptor 3 (TLR3) signaling has a protective role in innate immune response in cases of severe SARS-CoV infection. The FDA approved statins are considered TLR-MYD88 agonists, they keep or retains TLR-MYD88 level during hypoxia.164 Synchronously, an early and high dose of a statin might be an idea for treatment of MERS-CoV infections. However, statins may not be very effective for late-stage patients. Timely administration of statins may be crucial to surviving MERS-CoV infection.165 Importantly, a recent research study showed that angiotensin receptor blockers and statins upregulate ACE2, the tissue receptor for COVID-19; these effects participated in protection against acute respiratory disease syndrome and can decrease mortality rate.166 Statins are believed to reduce the risk of mortality in cases of influenza viral infection due to their anti-inflammatory and immunomodulatory effect.167 Moreover, rosuvastatin reduced cytokines TNF-a, IFN-g and Th-1 immune response during 72 h, it exerts rapid immunomodulatory effects.168 It is important to note that in all studies, there was no harm associated with statin therapy. Therefore, it is conceivable that patients admitted166 with viral respiratory illnesses including COVID-19 could derive a beneficial effect from the continuation of their statin therapy. Statins especially rosuvastatin are advised to be given to acute COVID-19 patients; they have a role in decreasing cardiovascular problems like the fatal myocardial infarction associated with COVID-19 infection.169
2.5 Rheumatoid arthritis drugs
Disease-Modifying Antirheumatic Drugs (DMARDs), Fig. 12 are a diverse collection of drugs, grouped according their use and convention, for treatment of rheumatoid arthritis (RA) and other related inflammatory diseases. DMARDs are immunosuppressive and immunomodulatory agents that have been found to improve symptoms, decrease joint damage, different types of pains and inflammations that affect, heart and blood vessels.170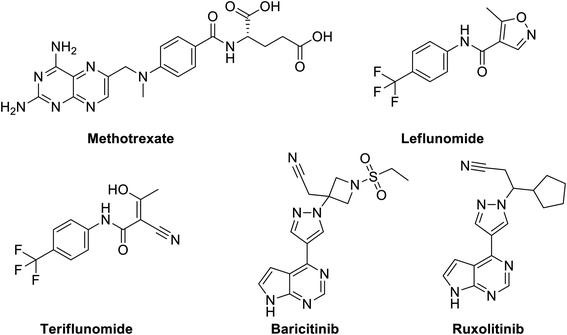 | ||
| Fig. 12 Structure of methotrexate, leflunomide, teriflunomide, baricitinib and ruxolitinib as examples of DMARDs. | ||
They are classified as either conventional DMARDs or biologic DMARDs. Commonly used conventional DMARDs therapeutic agents have fast onset with respect to their anti-inflammatory and analgesic effects but they neither remove the underlying cause of the disease nor protect against the functional disability.171 They include methotrexate (MTX), leflunomide, hydroxychloroquine, and sulfasalazine. Biologic DMARDs were introduced in the early 1990s and are usually prescribed after the failure of conventional DMARD therapy with advantages of more efficacy and fewer side effects.172 Biologic DMARDs are highly specific and target a specific pathway of the immune system against inflammation like inhibition of tumor necrosis factor, suppression of IL-1 and TNF-α, induction of apoptosis of inflammatory cells, by increasing chemotactic factors, inhibition of purine synthesis, pyrimidine metabolism or purine metabolism. They include TNF-alpha inhibitors (certolizumab, infliximab and etanercept), modified antibody (Abatacept), modified human interleukin 1 receptor antagonist protein (Anakinra), Janus associated kinase (JAK) inhibitors (baricitinib and tofacitinib) and monoclonal antibodies as interleukin-6 receptor blockers (e.g. tocilizumab, sarilumab, and rituximab).173
A number of classic DMARDs have demonstrated not only antitumor but unexpected antiviral activities such as methotrexate (MTX, Fig. 12) against mosquito-borne flaviviruses, such as dengue virus (DENV) and Zika virus (ZIKV) through dihydrofolate reductase (DHFR) inhibition mechanism leading to decreased viral replication.174 Other examples include leflunomide, Fig. 12, an isoxazole derivative, is nucleotide biosynthesis inhibitor, that is rapidly metabolized to its active form, and teriflunomide, which is a potent inhibitor of mitochondrial dihydroorotate dehydrogenase (DHODH), a key enzyme in the de novo biosynthesis of pyrimidine nucleoside triphosphates.175 Both leflunomide has demonstrated antiviral activity toward diverse DNA and RNA viruses, such as herpes simplex virus (HSV),176 human cytomegalovirus (HCMV),177 polyoma BK virus (BKV),178,179 human immunodeficiency virus (HIV),180 respiratory syncytial virus (RSV),181 and arenaviruses.182 Currently, they have not been tested against SARS-COV-2.
Currently, there are no specific antivirals or vaccines to treat SARS-CoV-2 infection. High-throughput screening (HTS) of various compound libraries against SARS-CoV-2 have been started and several biologic DMARDs were identified as possible targets in controlling SARS-COV-2 complications such as tocilizumab, sarilumab and baricitinib, Fig. 12.
Interleukin 6 (IL-6), a glycoprotein composed of 212 amino acids in humans, has pleiotropic effects on many cells, including B cells, T cells, hematopoietic stem cells, hepatocytes, megakaryocytes, osteoclasts, synoviocytes, keratinocytes, and possibly chondrocytes, and thus has a wide range of biological activity, including regulation of immune response, support of hematopoiesis, generation of acute phase reactions, and induction of inflammation and oncogenesis. Therapies involving blockade of IL-6 functions have constituted a new therapeutic strategy for some inflammatory and autoimmune diseases.183
Unlike conventional DMARDs, interleukin-6 antagonists (e.g. sarilumab and tocilizumab) have no direct or indirect antiviral activities; they block IL-6 a key cytokine that plays an important role in immune responses as proinflammatory cytokine that induces strong inflammatory responses. So, they may have a role in the severe deterioration of lung function and COVID-19 pneumonia.
Tocilizumab (TCZ), has been developed by Osaka University for the treatment of inflammatory and autoimmune disorders.184,185
TCZ was the first humanized IgG1 monoclonal antibodies acting as interleukin-6 (IL-6) receptor antagonist approved in Japan in 2008, in Europe in 2009, and FDA-approved in 2010 for the treatment of adult patients with moderately to severely active rheumatoid arthritis (RA) that didn't provide enough progress with other agents.186,187
TCZ can be administered intravenously or subcutaneously (SC) with similar efficacy, but convenience of SC formulation permits once-weekly self-administration.187 Extensive clinical experience has firmly established the short- and long-term efficacy and safety of tocilizumab (monotherapy or in combination with conventional DMARDs) in adults with early-stage and longer-duration established RA. In the clinical trial and real-world settings, tocilizumab monotherapy or combination therapy provided rapid and sustained improvements in clinical and radiographic outcomes and health-related quality of life. The safety profile of tocilizumab is consistent over time and, in general, is consistent with that of other immunomodulatory agents. It exhibits low immunogenicity.188
Tocilizumab has anticancer potency against non-small cell lung cancer cells via apoptosis induction as an agonistic IL-6R regulator and it is currently under evaluation in a multicenter clinical trial for large-cell lung carcinoma.189,190 In addition, tocilizumab completed phase II clinical trials for Pulmonary Arterial Hypertension (PAH) treatment.191
At present, this excellent safety and efficacy of tocilizumab encourages China's National Health Commission to include its use 8 mg kg−1/12 h (maximum of 800 mg per dose) in guidelines to treat coronavirus (COVID-19) patients either alone or in combination with favipiravir, a broad spectrum anti-viral agent. They report that the drug has been involved with a Chinese clinical trial involving 20 severe COVID-19 cases. 19 of them were discharged from the hospital within two weeks. Moreover, a 150-patient trial assessing tocilizumab is led by Hong Zhao of Peking University First Hospital and a 188-patient trial assessing tocilizumab alone is led by Dongsheng Wang of The First Affiliated Hospital of University of Science and Technology of China (Anhui Provincial Hospital).192
On 11 March 2020, in Italy, tocilizumab was the subject of an up-to-30-patient phase II trial designed to study the drug as a single 8 mg kg−1 dose in patients affected by severe pneumonia correlated to SARS-CoV2. The study by researchers at Università Politecnica delle Marche and Azienda Ospedaliera Ospedali Riuniti Marche Nord has two primary outcome measures: TCZ arrest in deterioration of pulmonary function, and improvement in pulmonary function. These signs of improvement prompting the Italian Pharmacological Agency (AIFA) to expand testing in 5 other hospitals.193
TCZ prevents overreaction of the immune system that has led to organ failure especially the lung and death in coronavirus patients. Roche and the WHO are each launching separate trials for its use in severe COVID-19 cases. Finally, Genentech (Roche) announced that the FDA has approved the initiation of a double-blind, randomized phase III clinical trial of the oncology supportive care drug tocilizumab (Actemra) for use in combination with standard care for the treatment of hospitalized adult patients with severe COVID-19 pneumonia.194
On February 15th, 2020, a month after the first reports of a novel coronavirus spreading in China. The artificial intelligence (AI) research groups and others have used AI software to find an already-approved drug that might limit the virus's ability to infect people.198 They used the first genomic sequence of the virus published in mid-January, and by January 29th, 2020 scientists suggested that the virus might enter human cells by binding to a cell-surface molecule called ACE2.199
The software pointed at the enzyme adaptor-associated protein kinase 1 (AAK1) as a possible target for the disease. AAK1 regulates endocytosis, the process that brings material into cells, which also is a common mode of viral infection. AI results select baricitinib, based on its affinity for the kinase and its toxicity amongst more than 378 known AAK1 inhibitors. Janus-associated kinase (JAK) inhibitor baricitinib is predicted to reduce the ability of the virus to infect lung cells by inhibition of ACE2-mediated endocytosis.198
Later, the researchers propose testing baricitinib in clinical trials against the virus as a dual antiviral and anti-inflammatory agent after confirming in vitro activities. Currently, ruxolitinib, Fig. 1, a drug that works by a similar mechanism, shows a promise in sever COVID-19-associated cytokine storm, where levels of cytokines were found to be significantly reduced. Also, It was found to be well-tolerated with low toxicities. It is subjected for phase III clinical trials for COVID-19.200
2.6 Miscellaneous drugs
Some other drug molecules or agents have a potential positive impact in management of COVID-infections. Research reports suggest that the use of antioxidants like ascorbic acid, N-acetylcysteine in combination with antiviral drugs can synergistically decrease the lethal effect of influenza viral infection.201 Another research study showed that combination of the influenza drug oseltamivir with N-acetylcysteine can increase host defense mechanism and decrease mortality rates. This occurs through reduction of oxidative stress associated with viral infection.202 Dietary supplements such as resveratrol, curcumin and sulphorafane were found to have cytoprotective effect on vital organs. This approach could be extrapolated to the lungs of infected patients with coronaviruses infections. A recent research study revealed that combination thalidomide and the selective COX-2 inhibitor, celecoxib, can improve the severe COVID-19 pneumonia through modulation of activated NF-KB that promote severe lung injury.203 The FDA-approved antiparasitic drug ivermectin (Fig. 13) is known in the literature by its broad-spectrum antiviral activity. Surprisingly, recent reports showed that this drug is a potent inhibitor of SARS-Cov-2. Ivermectin causes 5000× reduction in viral RNA during two days, it requires future clinical investigation in humans.2043 Conclusion
The war between scientists and viral infection is a continuous war. Finding specific potent drug with high efficacy and acceptable toxicity profile is a dream. It is the first time in the world to join scientists, politicians, diplomats, businessmen and the normal people all over the world to have the same target. Although the FDA approved chloroquine and hydroxychloroquine to be used in the emergency clinical management protocols used in CoV, this approval has some restrictions. The information about the wide use of these drugs is not enough. Some of the polymerase nucleoside/nucleotide inhibitors are considered promising agents. Favipiravir has selectivity on viral cells as it is viral RNA-dependent RNA polymerase inhibitor without effects on human cell lines; it has no effect on RNA or DNA in human cells. It is approved to be tested in clinical trials in cases of COVID-19 infections. Moreover, the combination between lopinavir and ritonavir is under investigation for clinical studies. The broad-spectrum antiviral effect of remdesivir against different RNA viruses in addition to its interference effect on CoV replication machinery recommend this drug for intensive studies to be COVID-19 drug.On the other hand, the ability of protease inhibitors to fight COVID-19 is under investigation. Inhibition of this enzyme is crucial for viral proteolytic and other essential vital activities. Lopinavir, ritonavir and nelfinavir are effective agents in COVID-19 infection but still need additional studies. Similarly, the serine protease inhibitors such as nafamostat and camostat are promising strategy in COVID-19 therapy. The important relatively save and potent lipid lowering statin, rosovastatin, has a good impact on lung pathological changes through decreasing Th2 and Th17 mediated cytokines. Statins are recommended to be useful in reducing the mortality rates in cases of influenza infections due to their anti-inflammatory and immunomodulatory effects. Some rheumatoid arthritis drugs may be beneficial in management of COVID-19 cases such as TNF-alpha inhibitors, interleukin 1 receptor antagonists; Janus associated kinases (JAK) in addition to monoclonal antibodies which block interleukin 6 receptors. Tocilizumab is under clinical investigation to treat hospitalized severe COVID-19 associated pneumonia. Also, baricitinib and ruxolitinib are suggested to induce ACE-2 mediated endocytosis that reduces ability of the virus to infect viral cells with their additional anti-inflammatory actions.
In the near future, scientists are invited to collect their efforts and share knowledge in redirecting the mentioned drugs in this review to have suitable, cheap drugs with the lowest possible toxicity profile.
Abbreviations
| COVID | Coronavirus disease |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| MERS | Middle East respiratory syndrome |
| PHEIC | Public health emergency of international concern |
| WHO | World Health Organization |
| HE | Hemagglutination-esterase |
| RBD | Receptor binding domain |
| ACE2 | Angiotensin converting enzyme 2 |
| TMPRSS2 | Transmembrane serine protease 2 |
| ORF1a | Open reading frame 1a |
| ORF1b | Open reading frame 1b |
| pp | Polyprotein |
| PLpro | Papain-like protease |
| 3CLpro | Chymotrypsin-like protease |
| RTC | Replicase–transcriptase complex |
| RdRp | RNA-dependent RNA polymerase |
| IFN | Interferon |
| APC | Antigen presenting cell |
| MHC | Major histocompatibility complex |
| CD8 | Cluster of differentiation |
| T cell = Tc | Cytotoxic lymphocyte |
| IgM | Immunoglobulin M |
| IgG | Immunoglobulin G |
| TMPRSS2 | Transmembrane protease, serine 2 |
| ICU | Intensive care unit |
| NK cells | Natural killer cell |
| RBD | Receptor-binding domain |
| VEGF | Vascular endothelial growth factor |
| HCMV | Human cytomegalovirus |
| HSV | Herpes simplex virus |
| BK | Polyomavirus |
| NIs | Nucleotide and nucleoside analogue inhibitors |
| FDA | Food and Drug Administration |
| TRP | Ribofuranosyl-5′-triphosphate |
| ATP | Adenosine triphosphate |
| GTP | Guanosine-5′-triphosphate |
| CT | Computerized tomography |
| IFNα2b | Interferon alpha-2b |
| nsp14 | Nonstructural protein 14 |
| HCV | Hepatitis C virus |
| RSV | Respiratory syncytial virus |
| NiV | Nipah virus |
| EBOV | Ebola virus |
| VEEV | Venezuelan equine encephalitis virus |
| IAV | Influenza A virus |
| CHIKV | Chikungunya virus |
| IBV | Influenza B virus |
| HCoV-NL63 | EC50: the concentration of a drug that gives half-maximal response |
| DAAs | Direct acting antivirals |
| ZIKV | Virus family Flaviviridae |
| TSG101 | Tumor susceptibility gene |
| RSV | Rosuvastatin |
| HMG-CoA | 3-Hydroxy-3-methyl-glutaryl-coenzyme A |
| UDP | Uridine diphosphate |
| TLR3 | Toll-like receptor 3 |
| MYD88 | Myeloid differentiation primary response 88 |
| Th1 | T helper cell |
| DMARD | Disease-modifying antirheumatic drugs |
| IL-1 | Interleukin-1 family |
| JAK | Janus associated kinase |
| DENV | Dengue virus |
| HCMV | Human cytomegalovirus |
| BKV | Polyoma BK virus |
| HTS | High-throughput screening |
| SC | Subcutaneously |
| RA | Rheumatoid arthritis |
| PAH BARDA | Pulmonary arterial hypertension; biomedical advanced research and development authority |
| AI | Artificial intelligence |
| AAK1 | Adaptor-associated protein kinase 1 |
| COX-2 | Cyclooxygenase 2 |
| NF-KB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| GIT | Gastrointestinal tract |
Conflicts of interest
Authors declare no conflict of interest.References
- C. C.-U. F. R. E. U. A. f. C. H. and U. S. F. D. A., https://www.fda.gov/news-events/pressannouncements/coronavirus-covid-19-update-fda-revokes-emergency-useauthorization-chloroquine-and., published 2020, accessed June 15, 2020, 2020.
- N. I. Nii-Trebi, BioMed Res. Int., 2017, 2017, 5245021 CrossRef PubMed.
- M. J. Walker, T. C. Barnett, J. D. McArthur, J. N. Cole, C. M. Gillen, A. Henningham, K. Sriprakash, M. L. Sanderson-Smith and V. Nizet, Clin. Microbiol. Rev., 2014, 27, 264–301 CrossRef PubMed.
- V. R. Racaniello, J. Clin. Invest., 2004, 113, 796–798 CrossRef CAS PubMed.
- C. Dye, Philos. Trans. R. Soc., B, 2014, 369, 20130426 CrossRef PubMed.
- F. Wu, S. Zhao, B. Yu, Y.-M. Chen, W. Wang, Z.-G. Song, Y. Hu, Z.-W. Tao, J.-H. Tian, Y.-Y. Pei, M.-L. Yuan, Y.-L. Zhang, F.-H. Dai, Y. Liu, Q.-M. Wang, J.-J. Zheng, L. Xu, E. C. Holmes and Y.-Z. Zhang, Nature, 2020, 579, 265–269 CrossRef CAS PubMed.
- World Health Organization, International health regulations (2005), World Health Organization, 2008 Search PubMed.
- World Health Organization, Naming the coronavirus disease (COVID-19) and the virus that causes it, 2020 Search PubMed.
- World Health Organization, World Health Organization best practices for the naming of new human infectious diseases, World Health Organization, 2015 Search PubMed.
- World Health Organization, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports, 2020.
- World Health Organization, Coronavirus disease (COVID-19) Situation Report – 153, June, 2020, p. 21 Search PubMed.
- S. L. Klein and K. L. Flanagan, Nat. Rev. Immunol., 2016, 16, 626–638 CrossRef CAS.
- Worldometer, Coronavirus Update (Live). 2020 Search PubMed.
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G. F. Gao and W. Tan, N. Engl. J. Med., 2020, 382, 727–733 CrossRef CAS.
- T. Pillaiyar, S. Meenakshisundaram and M. Manickam, Drug Discovery Today, 2020, 25, 668–688 CrossRef CAS PubMed.
- Y. Yin and R. G. Wunderink, Respirology, 2018, 23, 130–137 CrossRef PubMed.
- P. Zhou, X. L. Yang, X. G. Wang, B. Hu, L. Zhang, W. Zhang, H. R. Si, Y. Zhu, B. Li, C. L. Huang, H. D. Chen, J. Chen, Y. Luo, H. Guo, R. D. Jiang, M. Q. Liu, Y. Chen, X. R. Shen, X. Wang, X. S. Zheng, K. Zhao, Q. J. Chen, F. Deng, L. L. Liu, B. Yan, F. X. Zhan, Y. Y. Wang, G. F. Xiao and Z. L. Shi, Nature, 2020, 579, 270–273 CrossRef CAS PubMed.
- Z. Liu, X. Xiao, X. Wei, J. Li, J. Yang, H. Tan, J. Zhu, Q. Zhang and J. Wu, J. Med. Virol., 2020, 92, 595–601 CrossRef CAS PubMed.
- D. Wrapp, N. Wang, K. S. Corbett, J. A. Goldsmith, C.-L. Hsieh, O. Abiona, B. S. Graham and J. S. McLellan, Science, 2020, 367, 1260–1263 CrossRef CAS PubMed.
- I. Ghinai, T. D. McPherson, J. C. Hunter, H. L. Kirking, D. Christiansen, K. Joshi, R. Rubin, S. Morales-Estrada, S. R. Black, M. Pacilli, M. J. Fricchione, R. K. Chugh, K. A. Walblay, N. S. Ahmed, W. C. Stoecker, N. F. Hasan, D. P. Burdsall, H. E. Reese, M. Wallace, C. Wang, D. Moeller, J. Korpics, S. A. Novosad, I. Benowitz, M. W. Jacobs, V. S. Dasari, M. T. Patel, J. Kauerauf, E. M. Charles, N. O. Ezike, V. Chu, C. M. Midgley, M. A. Rolfes, S. I. Gerber, X. Lu, S. Lindstrom, J. R. Verani and J. E. Layden, Lancet, 2020, 395, 1137–1144 CrossRef CAS.
- C. Rothe, M. Schunk, P. Sothmann, G. Bretzel, G. Froeschl, C. Wallrauch, T. Zimmer, V. Thiel, C. Janke, W. Guggemos, M. Seilmaier, C. Drosten, P. Vollmar, K. Zwirglmaier, S. Zange, R. Wölfel and M. Hoelscher, N. Engl. J. Med., 2020, 382, 970–971 CrossRef PubMed.
- L. Zou, F. Ruan, M. Huang, L. Liang, H. Huang, Z. Hong, J. Yu, M. Kang, Y. Song, J. Xia, Q. Guo, T. Song, J. He, H. L. Yen, M. Peiris and J. Wu, N. Engl. J. Med., 2020, 382, 1177–1179 CrossRef PubMed.
- B. J. Bosch, R. van der Zee, C. A. de Haan and P. J. Rottier, J. Virol., 2003, 77, 8801–8811 CrossRef CAS PubMed.
- J. Armstrong, H. Niemann, S. Smeekens, P. Rottier and G. Warren, Nature, 1984, 308, 751–752 CrossRef CAS PubMed.
- B. Nal, C. Chan, F. Kien, L. Siu, J. Tse, K. Chu, J. Kam, I. Staropoli, B. Crescenzo-Chaigne, N. Escriou, S. van der Werf, K. Y. Yuen and R. Altmeyer, J. Gen. Virol., 2005, 86, 1423–1434 CrossRef CAS PubMed.
- P. Venkatagopalan, S. M. Daskalova, L. A. Lopez, K. A. Dolezal and B. G. Hogue, Virology, 2015, 478, 75–85 CrossRef CAS PubMed.
- A. Klausegger, B. Strobl, G. Regl, A. Kaser, W. Luytjes and R. Vlasak, J. Virol., 1999, 73, 3737–3743 CrossRef CAS.
- G. Simmons, P. Zmora, S. Gierer, A. Heurich and S. Pöhlmann, Antiviral Res., 2013, 100, 605–614 CrossRef CAS PubMed.
- A. Heurich, H. Hofmann-Winkler, S. Gierer, T. Liepold, O. Jahn and S. Pöhlmann, J. Virol., 2014, 88, 1293–1307 CrossRef PubMed.
- A. R. Fehr and S. Perlman, Methods Mol. Biol., 2015, 1282, 1–23 CrossRef CAS PubMed.
- F. Zhou, T. Yu, R. Du, G. Fan, Y. Liu, Z. Liu, J. Xiang, Y. Wang, B. Song, X. Gu, L. Guan, Y. Wei, H. Li, X. Wu, J. Xu, S. Tu, Y. Zhang, H. Chen and B. Cao, Lancet, 2020, 395, 1054–1062 CrossRef CAS.
- Y. Chen, K. Shan and W. Qian, Asians and Other Races Express Similar Levels of and Share the Same Genetic Polymorphisms of the SARS-CoV-2 Cell-Entry Receptor, Preprints, 2020, 2020020258, DOI:10.20944/preprints202002.0258.v1.
- D. Wang, B. Hu, C. Hu, F. Zhu, X. Liu, J. Zhang, B. Wang, H. Xiang, Z. Cheng, Y. Xiong, Y. Zhao, Y. Li, X. Wang and Z. Peng, JAMA, J. Am. Med. Assoc., 2020, 323, 1061–1069 CrossRef CAS PubMed.
- F. A. Rabi and M. S. Al Zoubi, Pathogens, 2020, 9, 231 CrossRef PubMed.
- J. Lessler, N. G. Reich, R. Brookmeyer, T. M. Perl, K. E. Nelson and D. A. Cummings, Lancet Infect. Dis., 2009, 9, 291–300 CrossRef.
- E. Prompetchara, C. Ketloy and T. Palaga, Asian Pac. J. Allergy Immunol., 2020, 38, 1–9 Search PubMed.
- Y. Shi, Y. Wang, C. Shao, J. Huang, J. Gan, X. Huang, E. Bucci, M. Piacentini, G. Ippolito and G. Melino, Cell Death Differ., 2020, 27, 1451–1454 CrossRef CAS PubMed.
- M. Liniger, A. Zuniga, A. Tamin, T. N. Azzouz-Morin, M. Knuchel, R. R. Marty, M. Wiegand, S. Weibel, D. Kelvin and P. A. Rota, Vaccine, 2008, 26, 2164–2174 CrossRef CAS PubMed.
- S. Salman and M. L. Salem, Med. Hypotheses, 2020, 140, 109689 CrossRef CAS PubMed.
- P. Baskar, G. Collins, B. Dorsey-Cooper, R. Pyle, J. Nagel, D. Dwyer, G. Dunston, C. Johnson, N. Kendig and E. Israel, Clin. Exp. Immunol., 1998, 111, 251 CrossRef CAS PubMed.
- S. Salman, M. S. Ahmed, A. M. Ibrahim, O. M. Mattar, H. El-Shirbiny, S. Sarsik, A. M. Afifi, R. M. Anis, N. A. Y. Agha and A. I. Abushouk, J. Am. Acad. Dermatol., 2019, 80, 922–930 CrossRef CAS PubMed.
- B. Gourbal, S. Pinaud, G. J. M. Beckers, J. W. M. Van Der Meer, U. Conrath and M. G. Netea, Immunol. Rev., 2018, 283, 21–40 CrossRef CAS PubMed.
- S. Yona, K.-W. Kim, Y. Wolf, A. Mildner, D. Varol, M. Breker, D. Strauss-Ayali, S. Viukov, M. Guilliams and A. Misharin, Immunity, 2013, 38, 79–91 CrossRef CAS PubMed.
- J. Kleinnijenhuis, J. Quintin, F. Preijers, L. A. Joosten, D. C. Ifrim, S. Saeed, C. Jacobs, J. van Loenhout, D. de Jong and H. G. Stunnenberg, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 17537–17542 CrossRef CAS PubMed.
- D.-L. Dixon, Children, 2015, 2, 289–304 CrossRef PubMed.
- E. Pandolfi, F. Gesualdo, C. Rizzo, E. Carloni, A. Villani, C. Concato, G. Linardos, L. Russo, B. Ferretti, I. Campagna and A. Tozzi, Frontiers in Pediatrics, 2019, 7, 152 CrossRef PubMed.
- S. Agarwal, W. Karmaus, S. Davis and V. Gangur, J. Hum. Lactation, 2011, 27, 171–186 CrossRef PubMed.
- K. M. Hunt, J. E. Williams, B. Shafii, M. K. Hunt, R. Behre, R. Ting, M. K. McGuire and M. A. McGuire, Breastfeed. Med., 2013, 8, 105–110 CrossRef PubMed.
- O. Ballard and A. L. Morrow, Pediatr. Clin. North Am., 2013, 60, 49–74 CrossRef PubMed.
- V. Surveillances, China CDC Weekly, 2020, 2, 113–122 Search PubMed.
- J. D. Berry, K. Hay, J. M. Rini, M. Yu, L. Wang, F. A. Plummer, C. R. Corbett and A. Andonov, mAbs, 2010, 2, 53–66 CrossRef PubMed.
- E. N. van den Brink, J. ter Meulen, F. Cox, M. A. Jongeneelen, A. Thijsse, M. Throsby, W. E. Marissen, P. M. Rood, A. B. Bakker and H. R. Gelderblom, J. Virol., 2005, 79, 1635–1644 CrossRef CAS PubMed.
- J. ter Meulen, E. N. van den Brink, L. L. Poon, W. E. Marissen, C. S. Leung, F. Cox, C. Y. Cheung, A. Q. Bakker, J. A. Bogaards, E. van Deventer, W. Preiser, H. W. Doerr, V. T. Chow, J. de Kruif, J. S. Peiris and J. Goudsmit, PLoS Med., 2006, 3, e237 CrossRef PubMed.
- J. ter Meulen, A. B. Bakker, E. N. van den Brink, G. J. Weverling, B. E. Martina, B. L. Haagmans, T. Kuiken, J. de Kruif, W. Preiser and W. Spaan, Lancet, 2004, 363, 2139–2141 CrossRef CAS.
- Z. Zhu, S. Chakraborti, Y. He, A. Roberts, T. Sheahan, X. Xiao, L. E. Hensley, P. Prabakaran, B. Rockx and I. A. Sidorov, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12123–12128 CrossRef CAS PubMed.
- C. Yihai and C. Yuguo, Bevacizumab in Severe or Critical Patients With COVID-19 Pneumonia, 2020, https://www.smartpatients.com/trials/NCT04275414 Search PubMed.
- National Institute of Allergy and Infectious Diseases, NIH Clinical Trial of Investigational Vaccine for COVID-19 Begins, 2020, https://www.niaid.nih.gov/news-events/nih-clinical-trial-investigational-vaccine-covid-19-begins Search PubMed.
- E. Kim, G. Erdos, S. Huang, T. W. Kenniston, S. C. Balmert, C. D. Carey, V. S. Raj, M. W. Epperly, W. B. Klimstra, B. L. Haagmans, E. Korkmaz, L. D. Falo Jr and A. Gambotto, EBioMedicine, 2020, 55, 102743 CrossRef PubMed.
- S. Jawhara, Int. J. Mol. Sci., 2020, 21, 2272 CrossRef PubMed.
- E. De Clercq and G. Li, Clin. Microbiol. Rev., 2016, 29, 695–747 CrossRef PubMed.
- A. A. Johnson, A. S. Ray, J. Hanes, Z. Suo, J. M. Colacino, K. S. Anderson and K. A. Johnson, J. Biol. Chem., 2001, 276, 40847–40857 CrossRef CAS PubMed.
- P. C. Jordan, S. K. Stevens and J. Deval, Antiviral Chem. Chemother., 2018, 26, 1–19 Search PubMed.
- T. P. Sheahan, A. C. Sims, R. L. Graham, V. D. Menachery, L. E. Gralinski, J. B. Case, S. R. Leist, K. Pyrc, J. Y. Feng and I. Trantcheva, Sci. Transl. Med., 2017, 9, eaal3653 CrossRef PubMed.
- E. C. Smith, H. Blanc, M. C. Surdel, M. Vignuzzi and M. R. Denison, PLoS Pathog., 2013, 9, e1003565 CrossRef CAS PubMed.
- M. Saijo, S. Morikawa, S. Fukushi, T. Mizutani, H. Hasegawa, N. Nagata, N. Iwata and I. Kurane, Antiviral Res., 2005, 66, 159–163 CrossRef CAS PubMed.
- E. J. Mifsud, F. G. Hayden and A. C. Hurt, Antiviral Res., 2019, 169, 104545 CrossRef CAS PubMed.
- L. MDVI and ClinicalTrials. gov, Phase 3 Efficacy and Safety Study of Favipiravir for Treatment of Uncomplicated Influenza in Adults - T705US316, National Library of Medicine (US), Bethesda, MD, 2015 Search PubMed.
- http://www.chinadaily.com.cn, 2020.
- Z. Jin, L. K. Smith, V. K. Rajwanshi, B. Kim and J. Deval, PLoS One, 2013, 8, e68347 CrossRef CAS PubMed.
- T. Baranovich, S.-S. Wong, J. Armstrong, H. Marjuki, R. J. Webby, R. G. Webster and E. A. Govorkova, J. Virol., 2013, 87, 3741–3751 CrossRef CAS.
- S. P. Gupta, Viral Polymerases: Structures, Functions and Roles as Antiviral Drug Targets, Academic Press, 2018 Search PubMed.
- J. G. Julander, D. F. Smee, J. D. Morrey and Y. Furuta, Antiviral Res., 2009, 82, 169–171 CrossRef CAS PubMed.
- L. Delang, N. Segura Guerrero, A. Tas, G. Quérat, B. Pastorino, M. Froeyen, K. Dallmeier, D. Jochmans, P. Herdewijn and F. Bello, J. Antimicrob. Chemother., 2014, 69, 2770–2784 CrossRef CAS PubMed.
- Y. Furuta, K. Takahashi, K. Shiraki, K. Sakamoto, D. F. Smee, D. L. Barnard, B. B. Gowen, J. G. Julander and J. D. Morrey, Antiviral Res., 2009, 82, 95–102 CrossRef CAS PubMed.
- E. De Clercq, Chem.–Asian J., 2019, 14, 3962–3968 CrossRef CAS.
- D. Sissoko, C. Laouenan, E. Folkesson, A. B. M'Lebing, A. H. Beavogui, S. Baize, A. M. Camara, P. Maes, S. Shepherd, C. Danel, S. Carazo, M. N. Conde, J. L. Gala, G. Colin, H. Savini, J. A. Bore, F. Le Marcis, F. R. Koundouno, F. Petitjean, M. C. Lamah, S. Diederich, A. Tounkara, G. Poelart, E. Berbain, J. M. Dindart, S. Duraffour, A. Lefevre, T. Leno, O. Peyrouset, L. Irenge, N. Bangoura, R. Palich, J. Hinzmann, A. Kraus, T. S. Barry, S. Berette, A. Bongono, M. S. Camara, V. Chanfreau Munoz, L. Doumbouya, H. Souley, P. M. Kighoma, F. R. Koundouno, L. Réné, C. M. Loua, V. Massala, K. Moumouni, C. Provost, N. Samake, C. Sekou, A. Soumah, I. Arnould, M. S. Komano, L. Gustin, C. Berutto, D. Camara, F. S. Camara, J. Colpaert, L. Delamou, L. Jansson, E. Kourouma, M. Loua, K. Malme, E. Manfrin, A. Maomou, A. Milinouno, S. Ombelet, A. Y. Sidiboun, I. Verreckt, P. Yombouno, A. Bocquin, C. Carbonnelle, T. Carmoi, P. Frange, S. Mely, V. K. Nguyen, D. Pannetier, A. M. Taburet, J. M. Treluyer, J. Kolie, R. Moh, M. C. Gonzalez, E. Kuisma, B. Liedigk, D. Ngabo, M. Rudolf, R. Thom, R. Kerber, M. Gabriel, A. Di Caro, R. Wölfel, J. Badir, M. Bentahir, Y. Deccache, C. Dumont, J. F. Durant, K. El Bakkouri, M. Gasasira Uwamahoro, B. Smits, N. Toufik, S. Van Cauwenberghe, K. Ezzedine, E. D'Ortenzio, L. Pizarro, A. Etienne, J. Guedj, A. Fizet, E. Barte de Sainte Fare, B. Murgue, T. Tran-Minh, C. Rapp, P. Piguet, M. Poncin, B. Draguez, T. Allaford Duverger, S. Barbe, G. Baret, I. Defourny, M. Carroll, H. Raoul, A. Augier, S. P. Eholie, Y. Yazdanpanah, C. Levy-Marchal, A. Antierrens, M. Van Herp, S. Günther, X. de Lamballerie, S. Keïta, F. Mentre, X. Anglaret and D. Malvy, PLoS Med., 2016, 13, e1001967 CrossRef PubMed.
- A. Maxmen, Nature, 2020, 578, 347–348 CrossRef CAS PubMed.
- M. Wang, R. Cao, L. Zhang, X. Yang, J. Liu, M. Xu, Z. Shi, Z. Hu, W. Zhong and G. Xiao, Cell Res., 2020, 30, 269–271 CrossRef CAS.
- Q. Cai, M. Yang, D. Liu, J. Chen, D. Shu, J. Xia, X. Liao, Y. Gu, Q. Cai and Y. Yang, Engineering, 2020 DOI:10.1016/j.eng.2020.03.007.
- L. Dong, S. Hu and J. Gao, Drug Discoveries Ther., 2020, 14, 58–60 CrossRef PubMed.
- E. Minskaia, T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard and J. Ziebuhr, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 5108–5113 CrossRef CAS PubMed.
- D. Falzarano, E. De Wit, C. Martellaro, J. Callison, V. J. Munster and H. Feldmann, Sci. Rep., 2013, 3, 1686 CrossRef PubMed.
- D. Falzarano, E. De Wit, A. L. Rasmussen, F. Feldmann, A. Okumura, D. P. Scott, D. Brining, T. Bushmaker, C. Martellaro and L. Baseler, Nat. Med., 2013, 19, 1313–1317 CrossRef CAS PubMed.
- J. A. Al-Tawfiq, H. Momattin, J. Dib and Z. A. Memish, Int. J. Infect. Dis., 2014, 20, 42–46 CrossRef CAS PubMed.
- A. S. Omrani, M. M. Saad, K. Baig, A. Bahloul, M. Abdul-Matin, A. Y. Alaidaroos, G. A. Almakhlafi, M. M. Albarrak, Z. A. Memish and A. M. Albarrak, Lancet Infect. Dis., 2014, 14, 1090–1095 CrossRef CAS PubMed.
- S. Shalhoub, F. Farahat, A. Al-Jiffri, R. Simhairi, O. Shamma, N. Siddiqi and A. Mushtaq, J. Antimicrob. Chemother., 2015, 70, 2129–2132 CrossRef CAS PubMed.
- A. Zumla, J. F. Chan, E. I. Azhar, D. S. Hui and K. Y. Yuen, Nat. Rev. Drug Discovery, 2016, 15, 327–347 CrossRef CAS.
- L. Zhang, B. Cao, Y. Wang, D. Wen, W. Liu, J. Wang, G. Fan, L. Ruan, B. Song, Y. Cai, M. Wei, X. Li, J. Xia, N. Chen, J. Xiang, T. Yu, T. Bai, X. Xie, C. Li, Y. Yuan, H. Chen, H. Li, H. Huang, S. Tu, F. Gong, Y. Liu, Y. Wei, C. Dong, F. Zhou, X. Gu, J. Xu, Z. Liu, Y. Zhang, H. Li, L. Shang, K. Wang, K. Li, X. Zhou, X. Dong, Z. Qu, S. Lu, X. Hu, S. Ruan, S. Luo, J. Wu, L. Peng, F. Cheng, L. Pan, J. Zou, C. Jia, J. Wang, X. Liu, S. Wang, X. Wu, Q. Ge, J. He, H. Zhan, F. Qiu, L. Guo, C. Huang, T. Jaki, F. G. Hayden, P. W. Horby, D. Zhang and C. Wang, N. Engl. J. Med., 2020, 382, 1787–1799 CrossRef PubMed.
- M. L. Agostini, E. L. Andres, A. C. Sims, R. L. Graham, T. P. Sheahan, X. Lu, E. C. Smith, J. B. Case, J. Y. Feng and R. Jordan, mBio, 2018, 9, e00221-0021–8 CrossRef.
- T. K. Warren, R. Jordan, M. K. Lo, A. S. Ray, R. L. Mackman, V. Soloveva, D. Siegel, M. Perron, R. Bannister and H. C. Hui, Nature, 2016, 531, 381–385 CrossRef CAS.
- M. K. Lo, R. Jordan, A. Arvey, J. Sudhamsu, P. Shrivastava-Ranjan, A. L. Hotard, M. Flint, L. K. McMullan, D. Siegel and M. O. Clarke, Sci. Rep., 2017, 7, 43395 CrossRef PubMed.
- E. P. Tchesnokov, J. Y. Feng, D. P. Porter and M. Götte, Viruses, 2019, 11, 326 CrossRef CAS PubMed.
- T. P. Sheahan, A. C. Sims, S. R. Leist, A. Schäfer, J. Won, A. J. Brown, S. A. Montgomery, A. Hogg, D. Babusis and M. O. Clarke, Nat. Commun., 2020, 11, 1–14 Search PubMed.
- M. L. Holshue, C. DeBolt, S. Lindquist, K. H. Lofy, J. Wiesman, H. Bruce, C. Spitters, K. Ericson, S. Wilkerson, A. Tural, G. Diaz, A. Cohn, L. Fox, A. Patel, S. I. Gerber, L. Kim, S. Tong, X. Lu, S. Lindstrom, M. A. Pallansch, W. C. Weldon, H. M. Biggs, T. M. Uyeki and S. K. Pillai, N. Engl. J. Med., 2020, 382, 929–936 CrossRef CAS PubMed.
- G. Li and E. De Clercq, Nat. Rev. Drug Discovery, 2020, 19, 149–150 CrossRef CAS PubMed.
- G. Griffiths, R. Fitzgerald, T. Jaki, A. Corkhill, E. Marwood, H. Reynolds, L. Stanton, S. Ewings, S. Condie, E. Wrixon, A. Norton, M. Radford, S. Yeats, J. Robertson, R. Darby-Dowman, L. Walker, S. Khoo and U. N. Community, Trials, 2020, 21, 544 CrossRef CAS PubMed.
- G. Ciliberto, R. Mancini and M. G. Paggi, J. Exp. Clin. Cancer Res., 2020, 3939, 8686 Search PubMed.
- J. Grein, N. Ohmagari, D. Shin, G. Diaz, E. Asperges, A. Castagna, T. Feldt, G. Green, M. L. Green, F. X. Lescure and E. Nicastri, N. Engl. J. Med., 2020, 382, 2327–2336 CrossRef CAS PubMed.
- Y. Wang, D. Zhang, G. Du, R. Du, J. Zhao, Y. Jin, S. Fu, L. Gao, Z. Cheng, Q. Lu, Y. Hu, G. Luo, K. Wang, Y. Lu, H. Li, S. Wang, S. Ruan, C. Yang, C. Mei, Y. Wang, D. Ding, F. Wu, X. Tang, X. Ye, Y. Ye, B. Liu, J. Yang, W. Yin, A. Wang, G. Fan, F. Zhou, Z. Liu, X. Gu, J. Xu, L. Shang, Y. Zhang, L. Cao, T. Guo, Y. Wan, H. Qin, Y. Jiang, T. Jaki, F. G. Hayden, P. W. Horby, B. Cao and C. Wang, Lancet, 2020, 395, 1569–1578 CrossRef CAS.
- G. M. Keating and A. Vaidya, Drugs, 2014, 74, 273–282 CrossRef CAS PubMed.
- A. A. Elfiky, Med. Chem., 2019, 15, 130–137 CrossRef CAS.
- D. Mani, A. Wadhwani and P. T. Krishnamurthy, J. Young Pharm., 2019, 11, 117 CrossRef CAS.
- N. Urakova, V. Kuznetsova, D. K. Crossman, A. Sokratian, D. B. Guthrie, A. A. Kolykhalov, M. A. Lockwood, M. G. Natchus, M. R. Crowley and G. R. Painter, J. Virol., 2018, 92, e01965-01917 Search PubMed.
- M. Ehteshami, S. Tao, K. Zandi, H.-M. Hsiao, Y. Jiang, E. Hammond, F. Amblard, O. O. Russell, A. Merits and R. F. Schinazi, Antimicrob. Agents Chemother., 2017, 61, e02395-02316 Search PubMed.
- J.-J. Yoon, M. Toots, S. Lee, M.-E. Lee, B. Ludeke, J. M. Luczo, K. Ganti, R. M. Cox, Z. M. Sticher and V. Edpuganti, Antimicrob. Agents Chemother., 2018, 62, e00766-00718 CrossRef.
- K. Pyrc, B. J. Bosch, B. Berkhout, M. F. Jebbink, R. Dijkman, P. Rottier and L. van der Hoek, Antimicrob. Agents Chemother., 2006, 50, 2000–2008 CrossRef CAS PubMed.
- L. J. Stuyver, T. Whitaker, T. R. McBrayer, B. I. Hernandez-Santiago, S. Lostia, P. M. Tharnish, M. Ramesh, C. K. Chu, R. Jordan and J. Shi, Antimicrob. Agents Chemother., 2003, 47, 244–254 CrossRef CAS PubMed.
- D. L. Barnard, V. D. Hubbard, J. Burton, D. F. Smee, J. D. Morrey, M. J. Otto and R. W. Sidwell, Antiviral Chem. Chemother., 2004, 15, 15–22 CrossRef CAS PubMed.
- B. Hannah, Drug Target Review, https://www.drugtargetreview.com/news/59567/eidd-2801-shows-efficacy-against-covid-19-in-human-cells-and-mice/, 2020 Search PubMed.
- R. Taylor, P. Kotian, T. Warren, R. Panchal, S. Bavari, J. Julander, S. Dobo, A. Rose, Y. El-Kattan, B. Taubenheim, Y. Babu and W. P. Sheridan, Journal of Infection and Public Health, 2016, 9, 220–226 CrossRef PubMed.
- M. Prajapat, P. Sarma, N. Shekhar, P. Avti, S. Sinha, H. Kaur, S. Kumar, A. Bhattacharyya, H. Kumar, S. Bansal and B. Medhi, Indian J. Pharmacol., 2020, 52, 56–65 CrossRef PubMed.
- D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020a, 368, 409–412 Search PubMed.
- J. Jacobs, V. Grum-Tokars, Y. Zhou, M. Turlington, S. A. Saldanha, P. Chase, A. Eggler, E. S. Dawson, Y. M. Baez-Santos and S. Tomar, J. Med. Chem., 2013, 56, 534–546 CrossRef CAS PubMed.
- M. R. Dayer, S. Taleb-Gassabi and M. S. Dayer, Arch. Clin. Infect. Dis., 2017, 12(4), 13823 Search PubMed.
- X. Liu and X. J. Wang, J. Genet. Genomics, 2020, 47, 119–121 CrossRef PubMed.
- T. R. Tong, Infect. Disord.: Drug Targets, 2009, 9, 223–245 CAS.
- A. K. Ghosh, H. L. Osswald and G. Prato, J. Med. Chem., 2016, 59, 5172–5208 CrossRef CAS PubMed.
- P. de Leuw and C. Stephan, GMS Infectious Diseases, 2017, 5 DOI:10.3205/id000034.
- S. Todd, C.-G. Anderson, D. J. Jolly and C. S. Craik, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 2000, 1477, 168–188 CrossRef CAS.
- J. Cinatl Jr, M. Michaelis, G. Hoever, W. Preiser and H. W. Doerr, Antiviral Res., 2005, 66, 81–97 CrossRef PubMed.
- U. Bacha, J. Barrila, A. Velazquez-Campoy, S. A. Leavitt and E. Freire, Biochemistry, 2004, 43, 4906–4912 CrossRef CAS PubMed.
- V. Nukoolkarn, V. S. Lee, M. Malaisree, O. Aruksakulwong and S. Hannongbua, J. Theor. Biol., 2008, 254, 861–867 CrossRef CAS PubMed.
- C. Chu, V. Cheng, I. Hung, M. Wong, K. Chan, K. Chan, R. Kao, L. Poon, C. Wong and Y. Guan, Thorax, 2004, 59, 252–256 CrossRef CAS PubMed.
- B. Cao, Y. Wang, D. Wen, W. Liu, J. Wang, G. Fan, L. Ruan, B. Song, Y. Cai, M. Wei, X. Li, J. Xia, N. Chen, J. Xiang, T. Yu, T. Bai, X. Xie, L. Zhang, C. Li, Y. Yuan, H. Chen, H. Li, H. Huang, S. Tu, F. Gong, Y. Liu, Y. Wei, C. Dong, F. Zhou, X. Gu, J. Xu, Z. Liu, Y. Zhang, H. Li, L. Shang, K. Wang, K. Li, X. Zhou, X. Dong, Z. Qu, S. Lu, X. Hu, S. Ruan, S. Luo, J. Wu, L. Peng, F. Cheng, L. Pan, J. Zou, C. Jia, J. Wang, X. Liu, S. Wang, X. Wu, Q. Ge, J. He, H. Zhan, F. Qiu, L. Guo, C. Huang, T. Jaki, F. G. Hayden, P. W. Horby, D. Zhang and C. Wang, N. Engl. J. Med., 2020, 382, 1787–1799 CrossRef PubMed.
- S. Chavez, B. Long, A. Koyfman and S. Y. Liang, Am. J. Emerg. Med., 2020 DOI:10.1016/j.ajem.2020.03.036.
- A. H. De Wilde, D. Falzarano, J. C. Zevenhoven-Dobbe, C. Beugeling, C. Fett, C. Martellaro, C. C. Posthuma, H. Feldmann, S. Perlman and E. J. Snijder, Virus Res., 2017, 228, 7–13 CrossRef CAS PubMed.
- N. Yamamoto, R. Yang, Y. Yoshinaka, S. Amari, T. Nakano, J. Cinatl, H. Rabenau, H. W. Doerr, G. Hunsmann and A. Otaka, Biochem. Biophys. Res. Commun., 2004, 318, 719–725 CrossRef CAS.
- H. Zhang, K. M. Saravanan, Y. Yang, M. T. Hossain, J. Li, X. Ren, Y. Pan and Y. Wei, Interdiscip. Sci.: Comput. Life Sci., 2020, 1–9 Search PubMed.
- L. Zhang, D. Lin, Y. Kusov, Y. Nian, Q. Ma, J. Wang, A. von Brunn, P. Leyssen, K. Lanko, J. Neyts, A. de Wilde, E. J. Snijder and H. Liu, J. Med. Chem., 2020, 63, 4562–4578 CrossRef CAS PubMed.
- A. Fischer and M. Sellner, Int. J. Mol. Sci., 2020, 21, 3626 CrossRef PubMed.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N. H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, Cell, 2020, 181, 271–280 CrossRef CAS PubMed.
- A. B. Patel and A. Verma, JAMA, J. Am. Med. Assoc., 2020, 323(23), 2386–2387 CrossRef CAS PubMed.
- I. Glowacka, S. Bertram, M. A. Müller, P. Allen, E. Soilleux, S. Pfefferle, I. Steffen, T. S. Tsegaye, Y. He and K. Gnirss, J. Virol., 2011, 85, 4122–4134 CrossRef PubMed.
- A. Shulla, T. Heald-Sargent, G. Subramanya, J. Zhao, S. Perlman and T. Gallagher, J. Virol., 2011, 85, 873–882 CrossRef CAS PubMed.
- M. Yamamoto, S. Matsuyama, X. Li, M. Takeda, Y. Kawaguchi, J.-i. Inoue and Z. Matsuda, Antimicrob. Agents Chemother., 2016, 60, 6532–6539 CrossRef CAS PubMed.
- Y. Kawaoka, M. Yamamoto, M. Kiso, Y. Sakai-Tagawa, K. Iwatsuki-Horimoto, M. Imai, M. Takeda, N. Kinoshita, N. Ohmagari, J. Gohda, K. Semba, Z. Matsuda, Y. Kawaguchi and J. Inoue, Viruses, 2020, 12, 629 CrossRef PubMed.
- M. A. A. Al-Bari, J. Antimicrob. Chemother., 2015, 70, 1608–1621 CrossRef.
- S. D'Alessandro, D. Scaccabarozzi, L. Signorini, F. Perego, D. P. Ilboudo, P. Ferrante and S. Delbue, Microorganisms, 2020, 8, 85 CrossRef PubMed.
- E. Keyaerts, L. Vijgen, P. Maes, J. Neyts and M. Van Ranst, Biochem. Biophys. Res. Commun., 2004, 323, 264–268 CrossRef CAS PubMed.
- E. Keyaerts, S. Li, L. Vijgen, E. Rysman, J. Verbeeck, M. Van Ranst and P. Maes, Antimicrob. Agents Chemother., 2009, 53, 3416–3421 CrossRef CAS PubMed.
- X. Yao, F. Ye, M. Zhang, C. Cui, B. Huang, P. Niu, X. Liu, L. Zhao, E. Dong, C. Song, S. Zhan, R. Lu, H. Li, W. Tan and D. Liu, Clin. Infect. Dis., 2020, ciaa237 CrossRef PubMed.
- M. J. Vincent, E. Bergeron, S. Benjannet, B. R. Erickson, P. E. Rollin, T. G. Ksiazek, N. G. Seidah and S. T. Nichol, Virol. J., 2005, 2, 69 CrossRef.
- A. Gomes, R. Ferraz, L. Ficker, M. S. Collins, C. Prudêncio, M. T. Cushion, C. Teixeira and P. Gomes, Antimicrob. Agents Chemother., 2018, 62, e00983-00918 Search PubMed.
- P. M. Njaria, J. Okombo, N. M. Njuguna and K. Chibale, Expert Opin. Ther. Pat., 2015, 25, 1003–1024 CrossRef CAS.
- OBI and E. Justice, Compositions and methods for treating warts associated with viral infections, Google Patents, WO 2014/008248 A2, 2014 Search PubMed.
- A. C. Kottkamp, E. De Jesus, R. Grande, J. A. Brown, A. R. Jacobs, J. K. Lim and K. A. Stapleford, J. Virol., 2019, 93, e00389-00319 Search PubMed.
- A. Farag, P. Wang, M. Ahmed and H. Sadek, ChemRxiv., 2020 DOI:10.26434/chemrxiv.12003930.v1.
- H.-H. Fan, L.-Q. Wang, W.-L. Liu, X.-P. An, Z.-D. Liu, X.-Q. He, L.-H. Song and Y.-G. Tong, Chin. Med. J., 2020, 133, 1051–1056 CrossRef PubMed.
- N. Salam, S. Mustafa, A. Hafiz, A. A. Chaudhary, F. Deeba and S. Parveen, BMC Public Health, 2018, 18, 710 CrossRef.
- J. Liu, R. Cao, M. Xu, X. Wang, H. Zhang, H. Hu, Y. Li, Z. Hu, W. Zhong and M. Wang, Cell Discovery, 2020, 6, 1–4 CrossRef.
- P. A. Gilormini, C. Lion, D. Vicogne, Y. Guérardel, F. Foulquier and C. Biot, J. Inherited Metab. Dis., 2018, 41, 515–523 CrossRef CAS.
- A. Savarino, L. Di Trani, I. Donatelli, R. Cauda and A. Cassone, Lancet Infect. Dis., 2006, 6, 67–69 CrossRef.
- J. Gao, Z. Tian and X. Yang, BioSci. Trends, 2020, 14, 72–73 CrossRef CAS PubMed.
- A. Cortegiani, G. Ingoglia, M. Ippolito, A. Giarratano and S. Einav, J. Crit. Care, 2020, 57, 279–283 CrossRef CAS PubMed.
- F. Touret and X. de Lamballerie, Antiviral Res., 2020, 177, 104762 CrossRef CAS PubMed.
- P. Gautret, J.-C. Lagier, P. Parola, L. Meddeb, M. Mailhe, B. Doudier, J. Courjon, V. Giordanengo, V. E. Vieira and H. T. Dupont, Int. J. Antimicrob. Agents, 2020, 105949 CrossRef.
- Z. Wu and J. M. McGoogan, JAMA, J. Am. Med. Assoc., 2020, 323(13), 1239–1242 CrossRef CAS PubMed.
- X. Yao, F. Ye, M. Zhang, C. Cui, B. Huang, P. Niu, X. Liu, L. Zhao, E. Dong and C. Song, Clin. Infect. Dis., 2020, ciaa237 CrossRef PubMed.
- M. R. Mehra, S. S. Desai, F. Ruschitzka and A. N. Patel, Lancet, 2020, 395, 102 CrossRef.
- J. Zhang, B. Xie and K. Hashimoto, Brain, Behav., Immun., 2020, 87, 59–73 CrossRef CAS PubMed.
- P. H. Jones, M. H. Davidson, E. A. Stein, H. E. Bays, J. M. McKenney, E. Miller, V. A. Cain, J. W. Blasetto and S. S. Group, Am. J. Cardiol., 2003, 92, 152–160 CrossRef CAS PubMed.
- S. Saadat, N. M. Roshan, M. R. Aslani and M. H. Boskabady, Cytokine, 2020, 128, 154993 CrossRef CAS PubMed.
- A. L. Totura, A. Whitmore, S. Agnihothram, A. Schäfer, M. G. Katze, M. T. Heise and R. S. Baric, mBio, 2015, 6, e00638-00615 Search PubMed.
- X. Yuan, Y. Deng, X. Guo, J. Shang, D. Zhu and H. Liu, Biochem. Biophys. Res. Commun., 2014, 446, 292–297 CrossRef CAS PubMed.
- S. Yuan, mBio, 2015, 6, e01120-01115 CrossRef PubMed.
- D. S. Fedson, S. M. Opal and O. M. Rordam, mBio, 2020, 11, e00398-00320 Search PubMed.
- M. L. Vandermeer, A. R. Thomas, L. Kamimoto, A. Reingold, K. Gershman, J. Meek, M. M. Farley, P. Ryan, R. Lynfield and J. Baumbach, J. Infect. Dis., 2012, 205, 13–19 CrossRef CAS PubMed.
- A. Link, T. Ayadhi, M. Böhm and G. Nickenig, Eur. Heart J., 2006, 27, 2945–2955 CrossRef CAS PubMed.
- C. S. Kow and S. S. Hasan, An International Journal of Medicine, 2020, 1–2 Search PubMed.
- J. Hamilton and V. Saravanan, Expert Opin. Pharmacother., 2002, 3, 845–856 CrossRef PubMed.
- M. G. Feely and J. R. O'Dell, Curr. Opin. Rheumatol., 2010, 22, 316–320 CrossRef CAS PubMed.
- P. Joshi and S. S. Dhaneshwar, Inflammation Allergy: Drug Targets, 2014, 13, 249–261 CrossRef CAS PubMed.
- M. Abbasi, M. J. Mousavi, S. Jamalzehi, R. Alimohammadi, M. H. Bezvan, H. Mohammadi and S. Aslani, J. Cell. Physiol., 2019, 234, 10018–10031 CrossRef CAS PubMed.
- S. Beck, Z. Zhu, M. F. Oliveira, D. M. Smith, J. N. Rich, J. A. Bernatchez and J. L. Siqueira-Neto, Viruses, 2019, 11, 338 CrossRef CAS PubMed.
- J. P. Davis, G. A. Cain, W. J. Pitts, R. L. Magolda and R. A. Copeland, Biochemistry, 1996, 35, 1270–1273 CrossRef CAS.
- D. A. Knight, A. Q. Hejmanowski, J. E. Dierksheide, J. W. Williams, A. S. Chong and W. J. Waldman, Transplantation, 2001, 71, 170–174 CrossRef CAS.
- D. L. Evers, X. Wang, S.-M. Huong, K. A. Andreoni and E.-S. Huang, Antiviral Res., 2005, 65, 1–12 CrossRef CAS PubMed.
- E. Bernhoff, G. D. Tylden, L. J. Kjerpeseth, T. J. Gutteberg, H. H. Hirsch and C. H. Rinaldo, J. Virol., 2010, 84, 2150–2156 CrossRef CAS.
- L. K. Jeffers-Francis, R. Burger-Calderon and J. Webster-Cyriaque, Antiviral Res., 2015, 118, 46–55 CrossRef CAS PubMed.
- E. Schläpfer, M. Fischer, P. Ott and R. F. Speck, AIDS, 2003, 17, 1613–1620 CrossRef.
- M. C. C. Dunn, D. A. Knight and W. J. Waldman, Antiviral Ther., 2011, 16, 309 CrossRef PubMed.
- C. S. Sepúlveda, C. C. García and E. B. Damonte, J. Med. Virol., 2018, 90, 819–827 CrossRef PubMed.
- K. Sato, M. Tsuchiya, J. Saldanha, Y. Koishihara, Y. Ohsugi, T. Kishimoto and M. M. Bendig, Cancer Res., 1993, 53, 851–856 CAS.
- T. Kishimoto, Clin. Rev. Allergy Immunol., 2005, 28, 177–185 CrossRef CAS.
- C. Ding and G. Jones, Rev. Recent Clin. Trials, 2006, 1, 193–200 CrossRef CAS PubMed.
- N. Nishimoto and T. Kishimoto, Handbook of experimental pharmacology, 2008, pp. 151–160 Search PubMed.
- A. Ogata, T. Hirano, Y. Hishitani and T. Tanaka, Clin. Med. Insights: Arthritis Musculoskeletal Disord., 2012, 5, CMAMD.S7371 Search PubMed.
- L. J. Scott, Drugs, 2017, 77, 1865–1879 CrossRef CAS PubMed.
- N. H. Kim, S. K. Kim, D. S. Kim, D. Zhang, J. A. Park, H. Yi, J. S. Kim and H. C. Shin, Oncol. Lett., 2015, 9, 2283–2288 CrossRef CAS PubMed.
- K. V. Korneev, K.-S. N. Atretkhany, M. S. Drutskaya, S. I. Grivennikov, D. V. Kuprash and S. A. Nedospasov, Cytokine, 2017, 89, 127–135 CrossRef CAS PubMed.
- K. W. Prins, T. Thenappan, E. K. Weir, R. Kalra, M. Pritzker and S. L. Archer, J. Am. Heart Assoc., 2019, 8, e011343 CAS.
- X. Xu, M. Han, T. Li, W. Sun and D. B. Wang, Proc. Natl. Acad. Sci. U. S. A., 2020, 5117(20), 10970–10975 CrossRef PubMed.
- X. Xu, M. Han, T. Li, W. Sun, D. Wang, B. Fu, Y. Zhou, X. Zheng, Y. Yang, X. Li, X. Zhang, A. Pan and H. Wei, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10970–10975 CrossRef CAS PubMed.
- G. Guaraldi, M. Meschiari, A. Cozzi-Lepri and J. Milic, Lancet Rheumatol, 2020 DOI:10.1016/S2665-9913(20)30210-1.
- E. G. Boyce, E. L. Rogan, D. Vyas, N. Prasad and Y. Mai, Ann. Pharmacother., 2018, 52, 780–791 CrossRef CAS PubMed.
- D. J Cennimo, 2020, https://www.medscape.com/answers/2500114-197456/what-is-the-role-of-the-il-6-inhibitor-sarilumab-kevzara-in-the-treatment-of-coronavirus-disease-2019-covid-19.
- A. Mogul, K. Corsi and L. McAuliffe, Ann. Pharmacother., 2019, 53, 947–953 CrossRef CAS.
- P. Richardson, I. Griffin, C. Tucker, D. Smith, O. Oechsle, A. Phelan and J. Stebbing, Lancet, 2020, 395, e30–e31 CrossRef CAS.
- Y. Zhao, Z. Zhao, Y. Wang, Y. Zhou, Y. Ma and W. Zuo, BioRxiv, 2020 DOI:10.1101/2020.01.26.919985.
- S. Yeleswaram, P. Smith, T. Burn, M. Covington, A. Juvekar, Y. Li, P. Squier and P. Langmuir, Clin. Immunol., 2020, 218, 108517 CrossRef CAS PubMed.
- N. Uchide and H. Toyoda, Molecules, 2011, 16, 2032–2052 CrossRef CAS PubMed.
- A. Garozzo, G. Tempera, D. Ungheri, R. Timpanaro and A. Castro, Int. J. Immunopathol. Pharmacol., 2007, 20, 349–354 CrossRef CAS PubMed.
- M. Hada, Chemotherapeutic Strategy with Synbiotics, Thalidomide and Celecoxib for severe COVID-19 Pneumonia. Association between microbiota, chronic inflammation and pneumonia, 2020, DOI:10.13140/RG.2.2.26979.91689.
- L. Caly, J. D. Druce, M. G. Catton, D. A. Jans and K. M. Wagstaff, Antiviral Res., 2020, 178, 104787 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |